

#microsky
#microsky
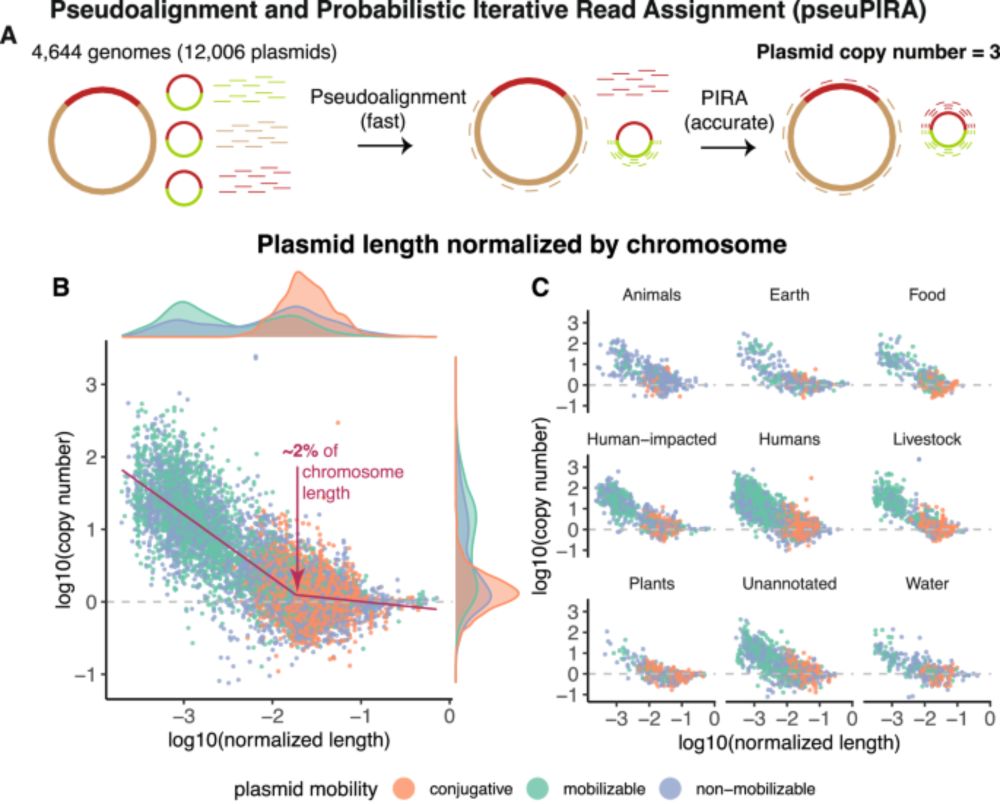
The Global Soil Plasmidome Resource: 98,728 soil plasmids from 6,860 samples.
Led by @mattlabguy.bsky.social and @apcamargo.bsky.social at @jgi.doe.gov @biosci.lbl.gov @berkeleylab.lbl.gov
www.nature.com/articles/s41...

The Global Soil Plasmidome Resource: 98,728 soil plasmids from 6,860 samples.
Led by @mattlabguy.bsky.social and @apcamargo.bsky.social at @jgi.doe.gov @biosci.lbl.gov @berkeleylab.lbl.gov
www.nature.com/articles/s41...
We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...

We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...
academic.oup.com/nar/advance-...

academic.oup.com/nar/advance-...
blast.ncbi.nlm.nih.gov/doc/blast-ne...
blast.ncbi.nlm.nih.gov/doc/blast-ne...
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

This problem has been studied extensively, and I recommend the survey of @camillemrcht.bsky.social, a great introduction to the subject
genome.cshlp.org/content/31/1...
Move & resize the ranges to see how that affects bedtools operations like merge and intersect in real time!
Move & resize the ranges to see how that affects bedtools operations like merge and intersect in real time!
Whenever you think you have a great idea in computational or evolutionary biology, it will already have been published by Eugene Koonin in the mid 90ies.

Whenever you think you have a great idea in computational or evolutionary biology, it will already have been published by Eugene Koonin in the mid 90ies.

myloasm-docs.github.io
myloasm-docs.github.io

Nunes et al. "Admixture’s impact on Brazilian population evolution and health" www.science.org/doi/10.1126/...
@hunemeier.bsky.social @macscastro.bsky.social 👏
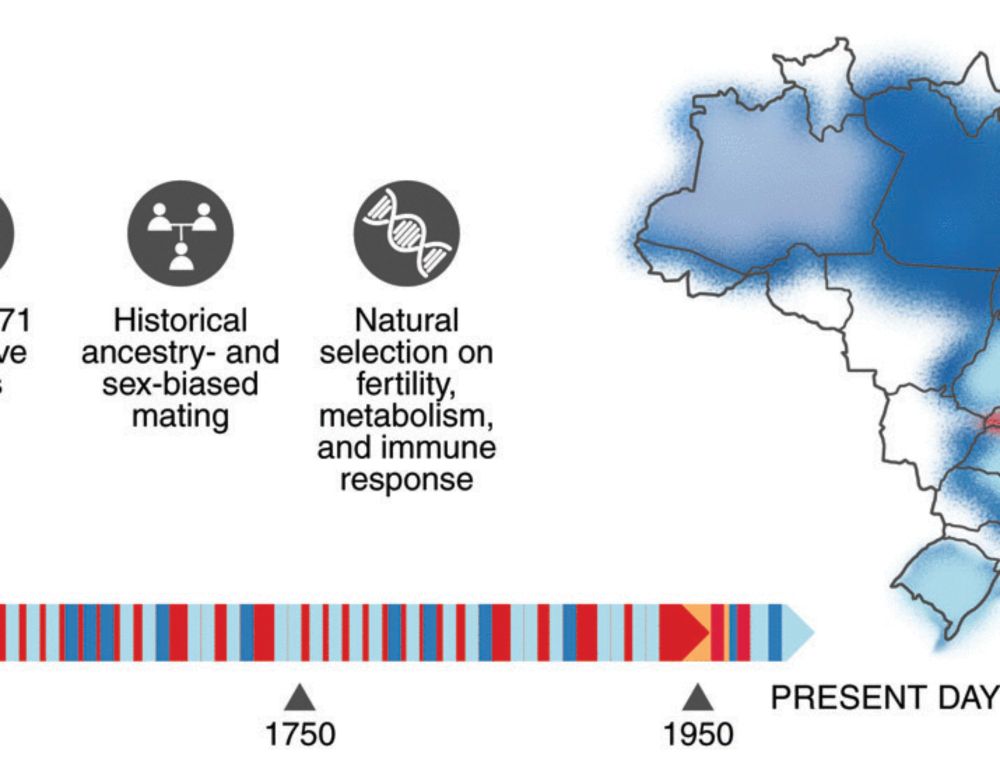
Nunes et al. "Admixture’s impact on Brazilian population evolution and health" www.science.org/doi/10.1126/...
@hunemeier.bsky.social @macscastro.bsky.social 👏
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
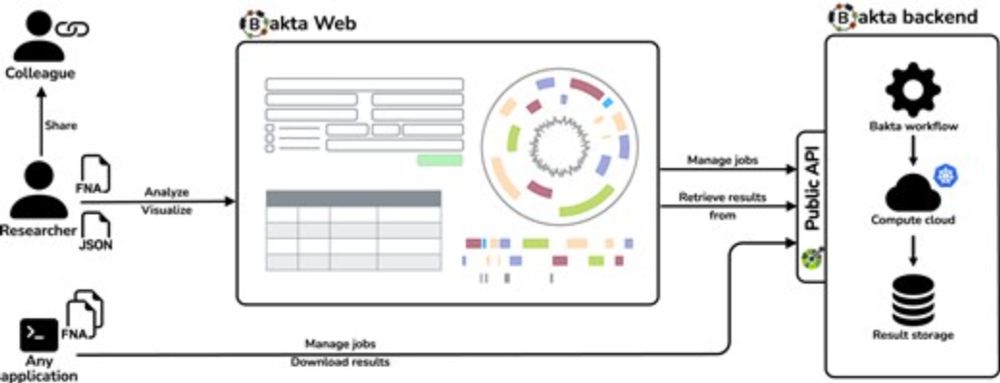
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
In particular these remarkably chimeric contigs are 😱, if rare....
Improving long read assemblers is definitely the space to be in when it comes to the future of metagenomics, as short reads won't be part of it 😉

In particular these remarkably chimeric contigs are 😱, if rare....
Improving long read assemblers is definitely the space to be in when it comes to the future of metagenomics, as short reads won't be part of it 😉

Martin Lercher and his team!
www.science.org/doi/10.1126/...

Martin Lercher and his team!
www.science.org/doi/10.1126/...
CoverM: Read alignment statistics for metagenomics academic.oup.com/bioinformati... #jcampubs

CoverM: Read alignment statistics for metagenomics academic.oup.com/bioinformati... #jcampubs

