Ben J Woodcroft
@benjwoodcroft.bsky.social
Yet another microbial bioinformatician, group leader, dad
github.com/wwood https://research.qut.edu.au/cmr/team/ben-woodcroft/
github.com/wwood https://research.qut.edu.au/cmr/team/ben-woodcroft/
Pinned

Out in @natbiotech.nature.com: Metagenome taxonomy profilers usually ignore unknown species. SingleM is an accurate profiler which doesn't, even detecting phyla with no MAGs. Profiles of 700,000 metagenomes at sandpiper.qut.edu.au. A 🧵
Reposted by Ben J Woodcroft
Excited to share a new preprint from the lab with @ryandhindsa.bsky.social ! www.biorxiv.org/content/10.1...
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/

July 22, 2025 at 9:59 PM
Excited to share a new preprint from the lab with @ryandhindsa.bsky.social ! www.biorxiv.org/content/10.1...
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/
Out in @natbiotech.nature.com: Metagenome taxonomy profilers usually ignore unknown species. SingleM is an accurate profiler which doesn't, even detecting phyla with no MAGs. Profiles of 700,000 metagenomes at sandpiper.qut.edu.au. A 🧵

July 16, 2025 at 9:59 PM
Out in @natbiotech.nature.com: Metagenome taxonomy profilers usually ignore unknown species. SingleM is an accurate profiler which doesn't, even detecting phyla with no MAGs. Profiles of 700,000 metagenomes at sandpiper.qut.edu.au. A 🧵
Reposted by Ben J Woodcroft
Very excited to share the first paper out of my Postdoc @CMR:
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social @jamesvolmer.bsky.social @benjwoodcroft.bsky.social
doi.org/10.1093/isme...
🧵1/7
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social @jamesvolmer.bsky.social @benjwoodcroft.bsky.social
doi.org/10.1093/isme...
🧵1/7

GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities
Abstract. Fluorescence in situ hybridisation (FISH) is a powerful tool for visualising the spatial organisation of microbial communities. However, traditio
doi.org
July 8, 2025 at 2:30 AM
Very excited to share the first paper out of my Postdoc @CMR:
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social @jamesvolmer.bsky.social @benjwoodcroft.bsky.social
doi.org/10.1093/isme...
🧵1/7
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social @jamesvolmer.bsky.social @benjwoodcroft.bsky.social
doi.org/10.1093/isme...
🧵1/7
Reposted by Ben J Woodcroft
Finally joined Bluesky! Follow along to stay connected with ABACBS. Keep an eye out in the coming days, we’ll be releasing more details for 2025 conference, including opening abstract submissions and announcing invited speakers!
www.abacbs.org/abacbs2025
www.abacbs.org/abacbs2025

ABACBS 2025 Conference
Adelaide, South Australia. Nov. 24-
www.abacbs.org
July 2, 2025 at 6:40 AM
Finally joined Bluesky! Follow along to stay connected with ABACBS. Keep an eye out in the coming days, we’ll be releasing more details for 2025 conference, including opening abstract submissions and announcing invited speakers!
www.abacbs.org/abacbs2025
www.abacbs.org/abacbs2025
Reposted by Ben J Woodcroft
I'm happy to announce the latest release of the GlobDB, available at globdb.org.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
home | GlobDB
globdb.org
June 10, 2025 at 11:20 AM
I'm happy to announce the latest release of the GlobDB, available at globdb.org.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
Reposted by Ben J Woodcroft
The Weather & Climate Livestream is officially LIVE from now until Sunday, June 1st! Our 💯 hours to #SaveAmericasForecasts starts today: wclivestream.com/watch/

May 28, 2025 at 5:09 PM
The Weather & Climate Livestream is officially LIVE from now until Sunday, June 1st! Our 💯 hours to #SaveAmericasForecasts starts today: wclivestream.com/watch/
Reposted by Ben J Woodcroft
Announcing myloasm, a new long-read (ONT R10/PacBio) metagenome assembler that I've been working on during my postdoc in the Heng Li lab (@lh3lh3.bsky.social).
myloasm-docs.github.io
myloasm-docs.github.io
myloasm - metagenomic assembly with (noisy) long reads
myloasm-docs.github.io
May 28, 2025 at 5:54 PM
Announcing myloasm, a new long-read (ONT R10/PacBio) metagenome assembler that I've been working on during my postdoc in the Heng Li lab (@lh3lh3.bsky.social).
myloasm-docs.github.io
myloasm-docs.github.io
@rossenzhao.bsky.social's shiny new phage profiling tool for finding (novel) phage in metagenome reads a la SingleM. New SingleM release also brings @ace-gtdb.bsky.social R226. Manuscript is draft, but working code is here pre-pre-publication. Feedback most welcome.

May 16, 2025 at 12:04 PM
@rossenzhao.bsky.social's shiny new phage profiling tool for finding (novel) phage in metagenome reads a la SingleM. New SingleM release also brings @ace-gtdb.bsky.social R226. Manuscript is draft, but working code is here pre-pre-publication. Feedback most welcome.
Reposted by Ben J Woodcroft
Ancient Host-Virus Gene Transfer Hints at a Diverse Pre-LECA Virosphere | Journal of Molecular Evolution https://link.springer.com/article/10.1007/s00239-025-10246-8#Bib1

Ancient Host-Virus Gene Transfer Hints at a Diverse Pre-LECA Virosphere - Journal of Molecular Evolution
The details surrounding the early evolution of eukaryotes and their viruses are largely unknown. Several key enzymes involved in DNA synthesis and transcription are shared between eukaryotes and large DNA viruses in the phylum Nucleocytoviricota, but the evolutionary relationships between these genes remain unclear. In particular, previous studies of eukaryotic DNA and RNA polymerases often show deep-branching clades of eukaryotes and viruses indicative of ancient gene exchange. Here, we performed updated phylogenetic analysis of eukaryotic and viral family B DNA polymerases, multimeric RNA polymerases, and mRNA-capping enzymes to explore their evolutionary relationships. Our results show that viral enzymes form clades that are typically adjacent to eukaryotes, suggesting that they originate prior to the emergence of the Last Eukaryotic Common Ancestor (LECA). The machinery for viral DNA replication, transcription, and mRNA capping are all key processes needed for the maintenance of virus factories, which are complex structures formed by many nucleocytoviruses during infection, indicating that viruses capable of making these structures are ancient. These findings hint at a diverse and complex pre-LECA virosphere and indicate that large DNA viruses may encode proteins that are relics of extinct proto-eukaryotic lineages.
link.springer.com
May 3, 2025 at 12:59 PM
Ancient Host-Virus Gene Transfer Hints at a Diverse Pre-LECA Virosphere | Journal of Molecular Evolution https://link.springer.com/article/10.1007/s00239-025-10246-8#Bib1
Reposted by Ben J Woodcroft
GTDB release 10 based on RefSeq 226 (R10-RS226) is live at gtdb.ecogenomic.org. This release covers 732,475 genomes (22% increase) and has 143,6141 species clusters (37% increase). Release notes at: forum.gtdb.ecogenomic.org/t/announcing.... Release statistics at: gtdb.ecogenomic.org/stats/r226.
GTDB - Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an initiative to establish a standardised microbial taxonomy based on genome phylogeny.
gtdb.ecogenomic.org
April 18, 2025 at 7:13 PM
GTDB release 10 based on RefSeq 226 (R10-RS226) is live at gtdb.ecogenomic.org. This release covers 732,475 genomes (22% increase) and has 143,6141 species clusters (37% increase). Release notes at: forum.gtdb.ecogenomic.org/t/announcing.... Release statistics at: gtdb.ecogenomic.org/stats/r226.
A 1.0 release for Sandpiper. 700,000 microbial community profiles (3x the last version, 4.7 Pbp metaG), searchable via the @ace-gtdb.bsky.social R226 taxonomy that just dropped. MetaGs are going exponential, but we are still nowhere near a MAG for all species. sandpiper.qut.edu.au #microsky 🧬🖥️ 1/2

April 17, 2025 at 11:06 AM
A 1.0 release for Sandpiper. 700,000 microbial community profiles (3x the last version, 4.7 Pbp metaG), searchable via the @ace-gtdb.bsky.social R226 taxonomy that just dropped. MetaGs are going exponential, but we are still nowhere near a MAG for all species. sandpiper.qut.edu.au #microsky 🧬🖥️ 1/2
We used Great Oxidation Event as a planet-sized "fossil" to add ancient dates to the Bacterial tree of life. @theconversation.com and @science.org articles show oxygen was used by non-cyanos before that cataclysm, surprisingly. theconversation.com/1-trillion-s... www.science.org/doi/10.1126/...

1 trillion species, 3 billion years: how we used AI to trace the evolution of bacteria on Earth
Until now, it’s been very hard for scientists to establish a detailed timeline of the early evolution of bacteria.
theconversation.com
April 4, 2025 at 2:12 AM
We used Great Oxidation Event as a planet-sized "fossil" to add ancient dates to the Bacterial tree of life. @theconversation.com and @science.org articles show oxygen was used by non-cyanos before that cataclysm, surprisingly. theconversation.com/1-trillion-s... www.science.org/doi/10.1126/...
Reposted by Ben J Woodcroft
We are looking for PhD students!
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
March 17, 2025 at 6:30 AM
We are looking for PhD students!
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
Reposted by Ben J Woodcroft
Check out this list of awesome #virome tools and be sure to add yours!
github.com/shandley/awe...
#phagesky #microsky
github.com/shandley/awe...
#phagesky #microsky

GitHub - shandley/awesome-virome: A listing of software, tools and databases useful for virome analysis
A listing of software, tools and databases useful for virome analysis - shandley/awesome-virome
github.com
March 12, 2025 at 8:46 PM
Check out this list of awesome #virome tools and be sure to add yours!
github.com/shandley/awe...
#phagesky #microsky
github.com/shandley/awe...
#phagesky #microsky
Reposted by Ben J Woodcroft
🦘✂️ Thanks to @cziscience.bsky.social , @wytamma.bsky.social and I are writing the next version of the Snippy bacterial variant calling pipeline. We want your input on what features s it will have. Please fill out this (longish) survey to help make Snippy great again!
forms.gle/YJP6WQjsk8KK...
forms.gle/YJP6WQjsk8KK...

Snippy: Microbial Variant Calling Community Survey
Help us drive the next wave of innovation in Single-Nucleotide Polymorphism (SNP) discovery and genome analysis by sharing your valuable experiences and insights. This in-depth survey aims at understa...
forms.gle
March 4, 2025 at 9:04 AM
🦘✂️ Thanks to @cziscience.bsky.social , @wytamma.bsky.social and I are writing the next version of the Snippy bacterial variant calling pipeline. We want your input on what features s it will have. Please fill out this (longish) survey to help make Snippy great again!
forms.gle/YJP6WQjsk8KK...
forms.gle/YJP6WQjsk8KK...
Wrapt for @iambrettb.bsky.social winning the student prize at @mgeaus.bsky.social - "Estimation of phage species trees using gene/species tree reconciliation". Hoping we can have an impact on viral phylogeny with these new methods, carefully applied.

February 18, 2025 at 6:07 AM
Wrapt for @iambrettb.bsky.social winning the student prize at @mgeaus.bsky.social - "Estimation of phage species trees using gene/species tree reconciliation". Hoping we can have an impact on viral phylogeny with these new methods, carefully applied.
Congrats to all here - a huge effort.
🚨🚨🚨 New pre-print alert: "Stable states in an unstable landscape: microbial resistance at the front line of climate change" doi.org/10.1101/2025...
Probably the most complex paper I have ever worked on. It took 4 (!) co-first authors to accomplish and literal years to decipher the story.
Probably the most complex paper I have ever worked on. It took 4 (!) co-first authors to accomplish and literal years to decipher the story.

Stable states in an unstable landscape: microbial resistance at the front line of climate change
Microbiome responses to warming may amplify or ameliorate terrestrial carbon loss and thus are a critical unknown in predicting climate outcomes. We quantified microbial warming response over seven ye...
doi.org
February 10, 2025 at 6:48 AM
Congrats to all here - a huge effort.
Reposted by Ben J Woodcroft
Excited to present my work on Bin Chicken at the next all-online MVIF conference! Come along if you are interested in using Bin Chicken for your own research.
It's Friday!
...and a new #MVIF program is out! 🤩
Registration here:
cassyni.com/s/mvif-36
⭐️ MicroTalks:
🇯🇵 Nodoka Chiba
🇮🇩 Patsy Tasyana Fitri
⭐️ Keynote:
🇯🇵 Ruixin Liu
⭐️ Selected talks:
🇦🇺 @aroneys.bsky.social
🇬🇧 Maria Kardakova
🇦🇺 Theo Portlock
...and a new #MVIF program is out! 🤩
Registration here:
cassyni.com/s/mvif-36
⭐️ MicroTalks:
🇯🇵 Nodoka Chiba
🇮🇩 Patsy Tasyana Fitri
⭐️ Keynote:
🇯🇵 Ruixin Liu
⭐️ Selected talks:
🇦🇺 @aroneys.bsky.social
🇬🇧 Maria Kardakova
🇦🇺 Theo Portlock

February 9, 2025 at 10:42 PM
Excited to present my work on Bin Chicken at the next all-online MVIF conference! Come along if you are interested in using Bin Chicken for your own research.
Will always be our first #rust project, a special place.
Excited to introduce the preprint for CoverM: the Swiss Army knife of coverage calculators for metagenomics! 🧬🖥️ DOI: doi.org/10.48550/arX...
@benjwoodcroft.bsky.social @viralinstruction.bsky.social @apcamargo.bsky.social
@benjwoodcroft.bsky.social @viralinstruction.bsky.social @apcamargo.bsky.social
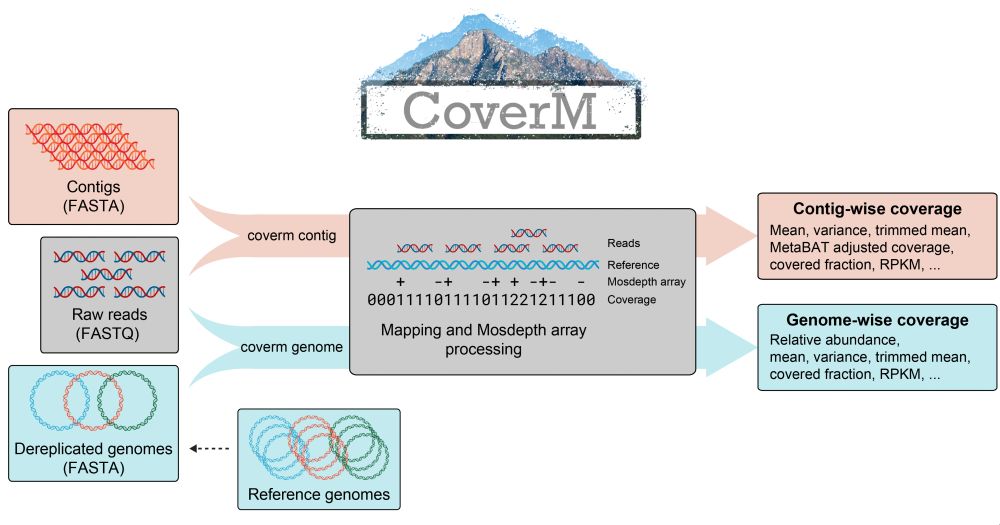
January 22, 2025 at 6:20 AM
Will always be our first #rust project, a special place.
Reposted by Ben J Woodcroft
Excited to share GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities
Fluorescence in situ hybridisation (FISH) is a powerful tool for visualising the spatial organisation of microbial communities. However, traditional FISH has several limitations, including limited p...
doi.org
December 19, 2024 at 5:17 PM
Excited to share GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
@sjmcilroy.bsky.social, @benjwoodcroft.bsky.social, @jamesvolmer.bsky.social
doi.org/10.21203/rs....
🧵1/6
GenomeFISH - a microbial microscopy method - make probes out of a genome (via SAG), and apply them back to the original sample - more specific, brighter, and broadly applicable than targeting 16S. Excellent from @pam-engelberts.bsky.social and others. www.researchsquare.com/article/rs-5...
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities
Fluorescence in situ hybridisation (FISH) is a powerful tool for visualising the spatial organisation of microbial communities. However, traditional FISH has several limitations, including limited p...
www.researchsquare.com
December 18, 2024 at 10:21 AM
GenomeFISH - a microbial microscopy method - make probes out of a genome (via SAG), and apply them back to the original sample - more specific, brighter, and broadly applicable than targeting 16S. Excellent from @pam-engelberts.bsky.social and others. www.researchsquare.com/article/rs-5...
Reposted by Ben J Woodcroft
I love Kingfisher! 🐦🐟
It's a fast and flexible program for procurement of sequence files (and their annotations) from public data sources, including the European Nucleotide Archive (ENA), NCBI SRA, Amazon AWS and Google Cloud. github.com/wwood/kingfi...
It's a fast and flexible program for procurement of sequence files (and their annotations) from public data sources, including the European Nucleotide Archive (ENA), NCBI SRA, Amazon AWS and Google Cloud. github.com/wwood/kingfi...

December 14, 2024 at 7:10 PM
I love Kingfisher! 🐦🐟
It's a fast and flexible program for procurement of sequence files (and their annotations) from public data sources, including the European Nucleotide Archive (ENA), NCBI SRA, Amazon AWS and Google Cloud. github.com/wwood/kingfi...
It's a fast and flexible program for procurement of sequence files (and their annotations) from public data sources, including the European Nucleotide Archive (ENA), NCBI SRA, Amazon AWS and Google Cloud. github.com/wwood/kingfi...
Reposted by Ben J Woodcroft
“Soil is likely home to 59% of life including everything from microbes to mammals, making it the singular most biodiverse habitat on Earth.”
Happy #WorldSoilDay!
www.pnas.org/doi/full/10....
Happy #WorldSoilDay!
www.pnas.org/doi/full/10....

December 5, 2024 at 7:47 PM
“Soil is likely home to 59% of life including everything from microbes to mammals, making it the singular most biodiverse habitat on Earth.”
Happy #WorldSoilDay!
www.pnas.org/doi/full/10....
Happy #WorldSoilDay!
www.pnas.org/doi/full/10....
Reposted by Ben J Woodcroft
Happy world soil day!
Soil of the year in Germany is the Renzina (Leptosol).
Rendzina is a shallow soil and common on carbonate-rich rocks. The name is Polish and describes the sound of a plough hitting the solid rock.
Here is my favourite photo I took in the alps.
Soil of the year in Germany is the Renzina (Leptosol).
Rendzina is a shallow soil and common on carbonate-rich rocks. The name is Polish and describes the sound of a plough hitting the solid rock.
Here is my favourite photo I took in the alps.

December 5, 2024 at 8:33 PM
Happy world soil day!
Soil of the year in Germany is the Renzina (Leptosol).
Rendzina is a shallow soil and common on carbonate-rich rocks. The name is Polish and describes the sound of a plough hitting the solid rock.
Here is my favourite photo I took in the alps.
Soil of the year in Germany is the Renzina (Leptosol).
Rendzina is a shallow soil and common on carbonate-rich rocks. The name is Polish and describes the sound of a plough hitting the solid rock.
Here is my favourite photo I took in the alps.
Reposted by Ben J Woodcroft
Enormous congratulations to Bob (Pok Man) Leung for receiving the Mollie Holman Award for Thesis Excellence. Bob was an exceptionally visionary, collaborative, and productive PhD student valued by all. He received an ARC DECRA Fellowship earlier this year to develop an independent program.


November 26, 2024 at 11:22 PM
Enormous congratulations to Bob (Pok Man) Leung for receiving the Mollie Holman Award for Thesis Excellence. Bob was an exceptionally visionary, collaborative, and productive PhD student valued by all. He received an ARC DECRA Fellowship earlier this year to develop an independent program.

