www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
www.nature.com/articles/s41...

www.nature.com/articles/s41...


🔗 DOI: www.biorxiv.org/content/10.6...

🔗 DOI: www.biorxiv.org/content/10.6...
rdcu.be/etIRy
This achievement marks a major step forward in the adoption and impact of the SeqCode within the scientific community.
Thank you to @ebi.embl.org for support with setting up these links!
Thank you to @ebi.embl.org for support with setting up these links!
⬇️
academic.oup.com/nar/advance-...

⬇️
academic.oup.com/nar/advance-...


The findings expand our understanding of how alternative genetic codes evolve and hint at new molecular tools for biotechnology applications. https://scim.ag/4omApQ7

The findings expand our understanding of how alternative genetic codes evolve and hint at new molecular tools for biotechnology applications. https://scim.ag/4omApQ7
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Join #MVIF 44 – we will discuss diverse #microbiome topics:
human urinary, gut, oral and nasal microbiomes,
extreme environments, and
a new sequence alignment tool.
...and a new #MVIF program is out! 🤩
Free registration: cassyni.com/s/mvif-44
⭐️ Highlights:
🇺🇸 Vanessa Hale
🇰🇷 Jun Hyung Cha
⭐️ Keynote:
🇺🇸 Katherine Lemon @kathlemon.bsky.social
⭐️ Talks:
🇺🇸 Meenakshi Chakraborty
🇨🇳 Wei Shen @shenwei356.bsky.social
🇺🇸 Johanna Gutleben

Join #MVIF 44 – we will discuss diverse #microbiome topics:
human urinary, gut, oral and nasal microbiomes,
extreme environments, and
a new sequence alignment tool.
academic.oup.com/nar/advance-...

academic.oup.com/nar/advance-...
Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
Explore the full study: https://doi.org/10.1093/bioadv/vbaf280

Explore the full study: https://doi.org/10.1093/bioadv/vbaf280
We’re thrilled to share that Gene Tyson, @benjwoodcroft.bsky.social and @luispedrocoelho.bsky.social have once again been named Highly Cited Researchers for 2025 by Clarivate!
#HighlyCited2025
We’re thrilled to share that Gene Tyson, @benjwoodcroft.bsky.social and @luispedrocoelho.bsky.social have once again been named Highly Cited Researchers for 2025 by Clarivate!
#HighlyCited2025
doi.org/10.1093/bioa...
The GlobDB is the largest species dereplicated genome database currently available, containing 306,260 species representatives.
More information on globdb.org 1/5
🖥️🧬🦠

doi.org/10.1093/bioa...
The GlobDB is the largest species dereplicated genome database currently available, containing 306,260 species representatives.
More information on globdb.org 1/5
🖥️🧬🦠
proGenomes4: providing 2 million accurately and consistently annotated high-quality prokaryotic genomes: academic.oup.com/nar/article/...

proGenomes4: providing 2 million accurately and consistently annotated high-quality prokaryotic genomes: academic.oup.com/nar/article/...
The Global Soil Plasmidome Resource: 98,728 soil plasmids from 6,860 samples.
Led by @mattlabguy.bsky.social and @apcamargo.bsky.social at @jgi.doe.gov @biosci.lbl.gov @berkeleylab.lbl.gov
www.nature.com/articles/s41...

The Global Soil Plasmidome Resource: 98,728 soil plasmids from 6,860 samples.
Led by @mattlabguy.bsky.social and @apcamargo.bsky.social at @jgi.doe.gov @biosci.lbl.gov @berkeleylab.lbl.gov
www.nature.com/articles/s41...
Microbial fraction is now prokaryotic fraction, easier to specify input genomes.
Microbial fraction is now prokaryotic fraction, easier to specify input genomes.
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
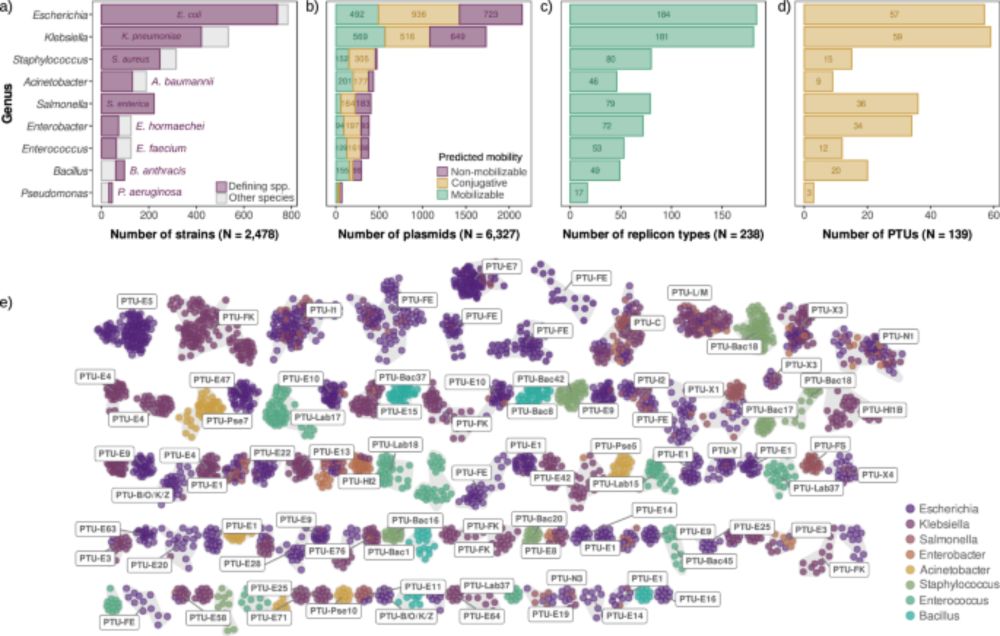
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6

doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6
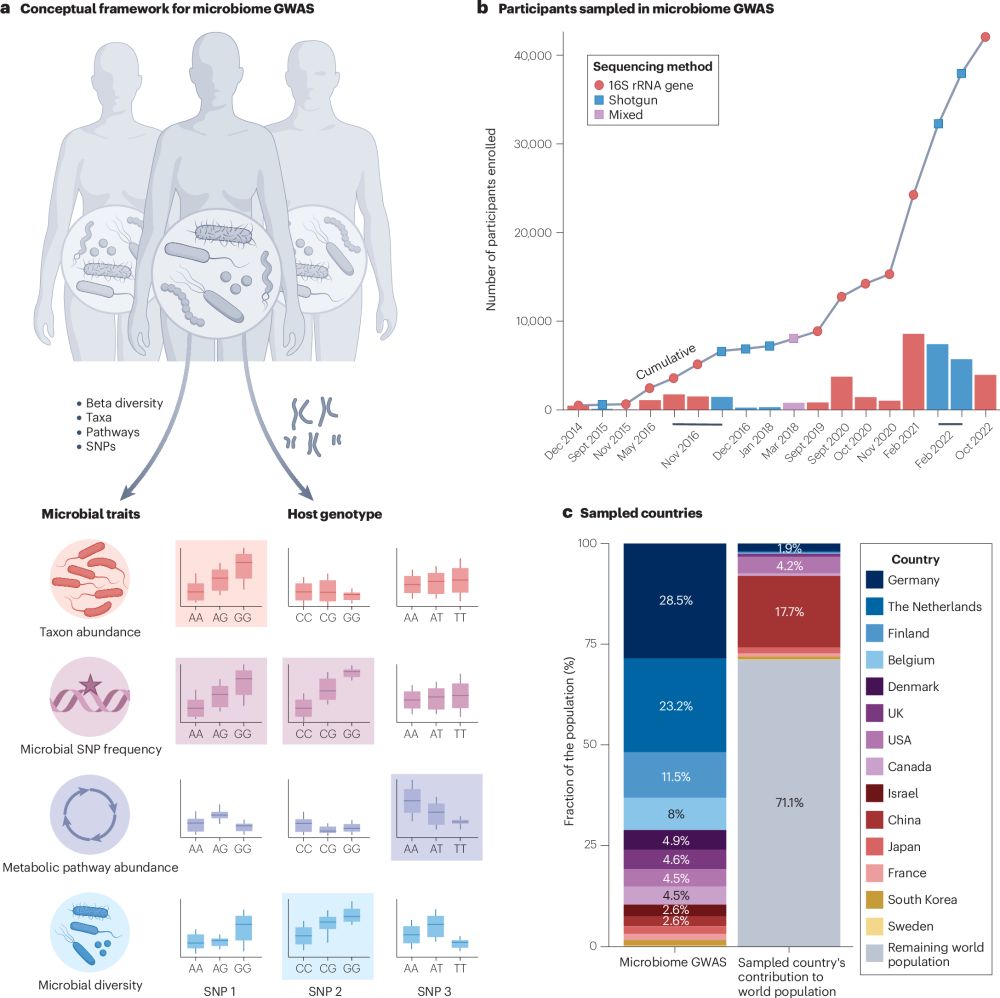
www.cell.com/trends/micro...

www.cell.com/trends/micro...

