This serves as a basis for predicted spectral libraries, reducing the search space 10x-20x. www.biorxiv.org/content/10.1...

This serves as a basis for predicted spectral libraries, reducing the search space 10x-20x. www.biorxiv.org/content/10.1...
github.com/FossatiLab/s...

github.com/FossatiLab/s...
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...

We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...
Tracing the arc of conceptual milestones in life sciences over the past century, centered around major discoveries driven by evolving technologies.
We are excited about the progress & the research enabled by tech from @ParallelSqTech.
youtu.be/SrzoLGBVTsY?...

Tracing the arc of conceptual milestones in life sciences over the past century, centered around major discoveries driven by evolving technologies.
We are excited about the progress & the research enabled by tech from @ParallelSqTech.
youtu.be/SrzoLGBVTsY?...
16 fully funded PhD positions are open in ProtAIomics Doctoral Network!
Join 16 labs across Europe to advance AI-powered proteomics and drive discoveries in health & biotechnology.
👉 Apply here: www.protaiomics.eu
#Proteomics #ArtificialIntelligence #MSCA #PhDOpportunities

Preprint: www.biorxiv.org/content/10.1...
@patiskowronek.bsky.social explains👇
📄 Preprint: doi.org/10.1101/2025... 1/🧵

Preprint: www.biorxiv.org/content/10.1...
@patiskowronek.bsky.social explains👇
👉 Improves imputation accuracy, normalization, and differential expression detection
📝https://www.biorxiv.org/content/10.1101/2025.10.02.679746v1
👉 Improves imputation accuracy, normalization, and differential expression detection
📝https://www.biorxiv.org/content/10.1101/2025.10.02.679746v1
They '...systematically screened 200 million human protein pairs and predicted 17,849 interactions with anexpected precision of 90%, of which 3,631 interactions were not identified in previous experimental screens.'
www.science.org/doi/epdf/10....

They '...systematically screened 200 million human protein pairs and predicted 17,849 interactions with anexpected precision of 90%, of which 3,631 interactions were not identified in previous experimental screens.'
www.science.org/doi/epdf/10....
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️

📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
#IchBinHanna #IchBinReyhan
library.oapen.org/bitstream/ha...
en.gdch.de/network-stru...
#ChemSky #ECR #EarlyCareerResearcher
Apply for our Meeting Award to get:
✅ 500 € grant for #Biochemistry2026
✅ 1 year of free @gdchbiochem.bsky.social membership
Apply now: en.gdch.de/network-s...
Share with a #ECR who needs to see this! 👇
#ChemSky #ChemBio #AcademicTwitter #PhD @gdch.de

en.gdch.de/network-stru...
#ChemSky #ECR #EarlyCareerResearcher
➡️ www.uni-wuerzburg.de/en/news-and-...

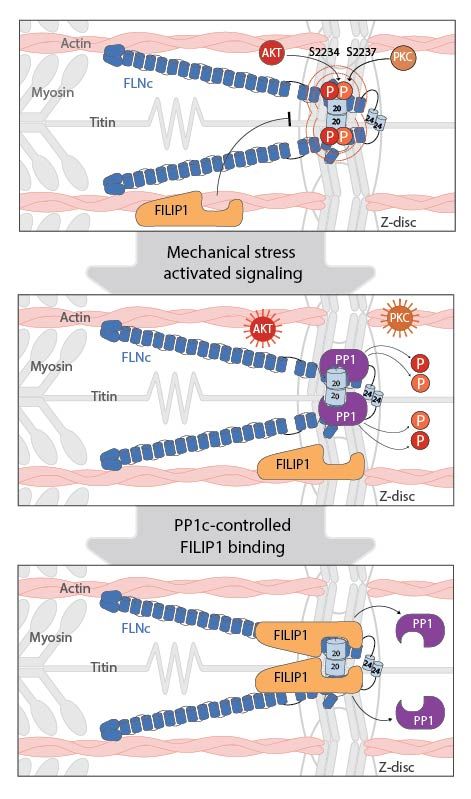
www.science.org/doi/10.1126/...
(1/4)
www.science.org/doi/10.1126/...
(1/4)
---
#proteomics #prot-paper
---
#proteomics #prot-paper
"...yielding ~15,000 and ~30,000 additional precursors..."
eDR: www.biorxiv.org/content/10.1...
MAP-MS: www.biorxiv.org/content/10.1...
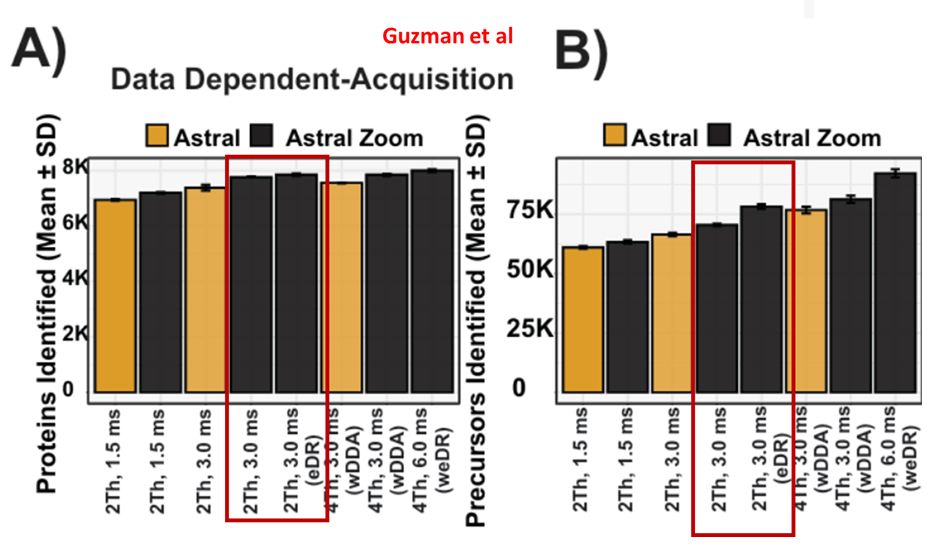
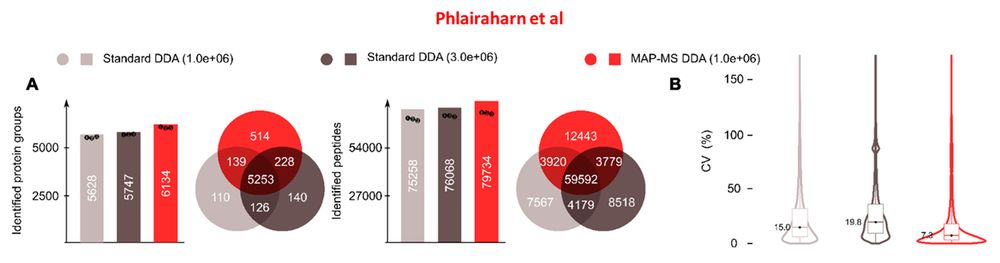
"...yielding ~15,000 and ~30,000 additional precursors..."
eDR: www.biorxiv.org/content/10.1...
MAP-MS: www.biorxiv.org/content/10.1...
Thanks @diegofgar.bsky.social for sharing!
#teammassspec
#AcademicSky
#ChemSky
Feel free to contact me or @sinzandrea.bsky.social
#Postdoc #MassSpec #Proteomics #Hiring




Thanks @diegofgar.bsky.social for sharing!
#teammassspec
#AcademicSky
#ChemSky

💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC
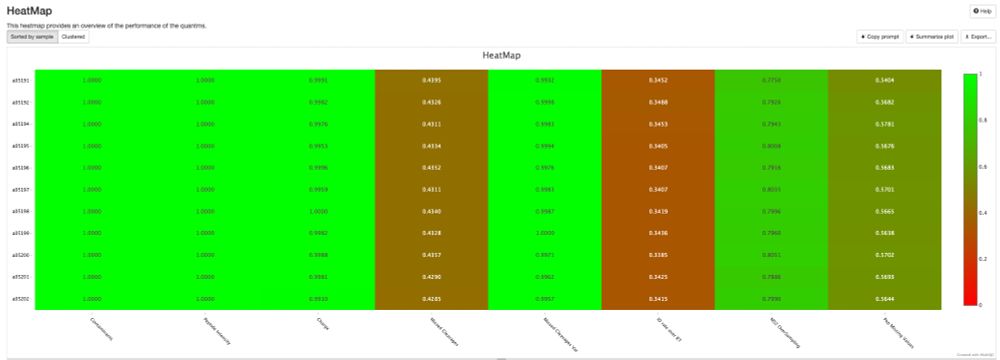
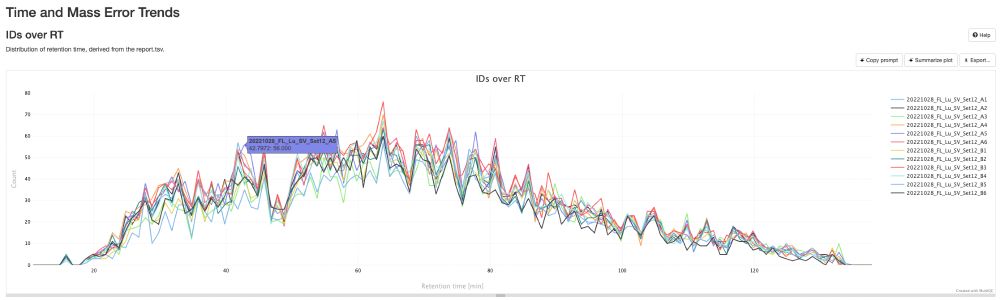
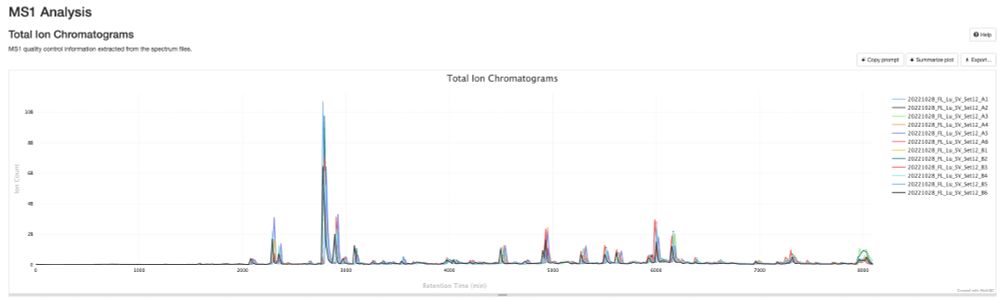
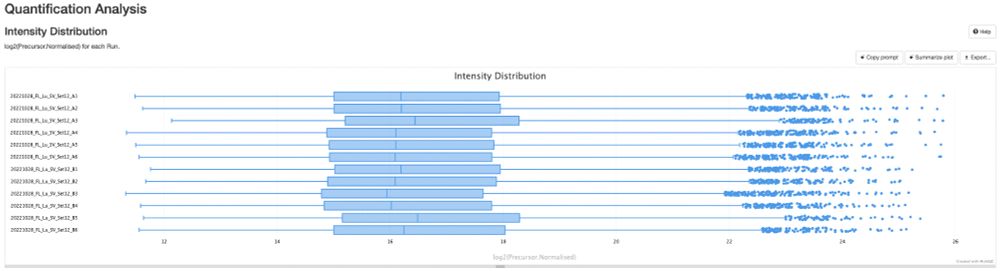
💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC
github.com/pwilmart/qua...
github.com/pwilmart/qua...
After more than 4.5 fantastic years at @embl.org, I am excited to announce that from 1 August 2025, I’ll be starting my own research group at Physikalisch-Technische Bundesanstalt (PTB) in Braunschweig, Germany — as part of the Biochemistry Department [1/n]
After more than 4.5 fantastic years at @embl.org, I am excited to announce that from 1 August 2025, I’ll be starting my own research group at Physikalisch-Technische Bundesanstalt (PTB) in Braunschweig, Germany — as part of the Biochemistry Department [1/n]
📅 Registration closes: July 7
📍 Utrecht, July 21–25
💻 Hands-on training in MaxQuant, Perseus & mass-spectrometry-based proteomics
🔗 Sign up now: eventbrite.de/e/maxquant-s...
🌐 Details: maxquant.org/summer_school/
#TeamMassSpec

📅 Registration closes: July 7
📍 Utrecht, July 21–25
💻 Hands-on training in MaxQuant, Perseus & mass-spectrometry-based proteomics
🔗 Sign up now: eventbrite.de/e/maxquant-s...
🌐 Details: maxquant.org/summer_school/
#TeamMassSpec

