

I'll finish my PhD work within the next few month and started to sort out where to continue doing exciting science. Any recommendations & suggestions are welcome🙏 More information on my scientific background and ideas for the future below 🧵 reskeet very much appreciated!

pubs.acs.org/doi/10.1021/...
github.com/snijderlab/a...
github.com/jspaezp/rust...

pubs.acs.org/doi/10.1021/...
github.com/snijderlab/a...
github.com/jspaezp/rust...


www.nature.com/articles/s41...

www.nature.com/articles/s41...

Read more here: www.biorxiv.org/content/10.1101/2025.08.30.673231v1

Read more here: www.biorxiv.org/content/10.1101/2025.08.30.673231v1
➡️ www.uni-wuerzburg.de/en/news-and-...

---
#proteomics #prot-paper

---
#proteomics #prot-paper


🧪 #relentlessdiscovery @rjdlab.bsky.social

🧪 #relentlessdiscovery @rjdlab.bsky.social
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec


Der Spitzenforscher wird glorifiziert, die notwendige Vorarbeit die von Dutzenden anderen Forschenden geleistet wird, wird ausgeblendet und herabgewürdigt
Der Spitzenforscher wird glorifiziert, die notwendige Vorarbeit die von Dutzenden anderen Forschenden geleistet wird, wird ausgeblendet und herabgewürdigt
„Für eine Dokumentation für den Öffentlich-rechtlichen Rundfunk suchen wir Wissenschaftler*innen ohne Lebenszeitprofessur, die bereit wären, mit uns über ihre Erfahrungen mit dem dt. Wissenschaftssystem zu sprechen:
(1/3)
„Für eine Dokumentation für den Öffentlich-rechtlichen Rundfunk suchen wir Wissenschaftler*innen ohne Lebenszeitprofessur, die bereit wären, mit uns über ihre Erfahrungen mit dem dt. Wissenschaftssystem zu sprechen:
(1/3)
www.nature.com/articles/s41...
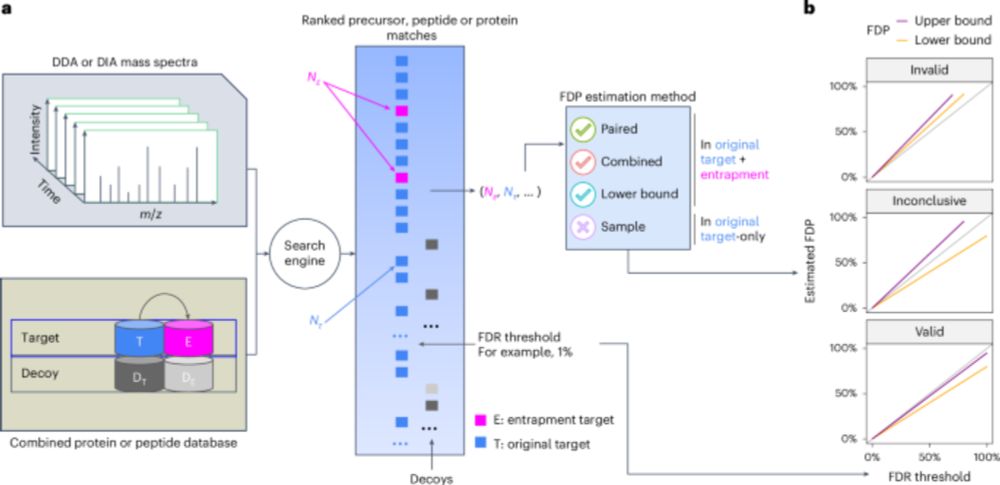

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.embopress.org/doi/full/10....

www.embopress.org/doi/full/10....
We demonstrate 27-plex DIA enabling over 500 samples / day.
We project that timePlex will enable throughputs exceeding 1,000 samples / day.
www.biorxiv.org/content/10.1...
We demonstrate 27-plex DIA enabling over 500 samples / day.
We project that timePlex will enable throughputs exceeding 1,000 samples / day.
www.biorxiv.org/content/10.1...

