Website: https://darusso.de/

Major highlight: 85% Synechococcus proteome coverage in a single shotgun experiment.
Out now in Plant Physiology!
doi.org/10.1093/plph...





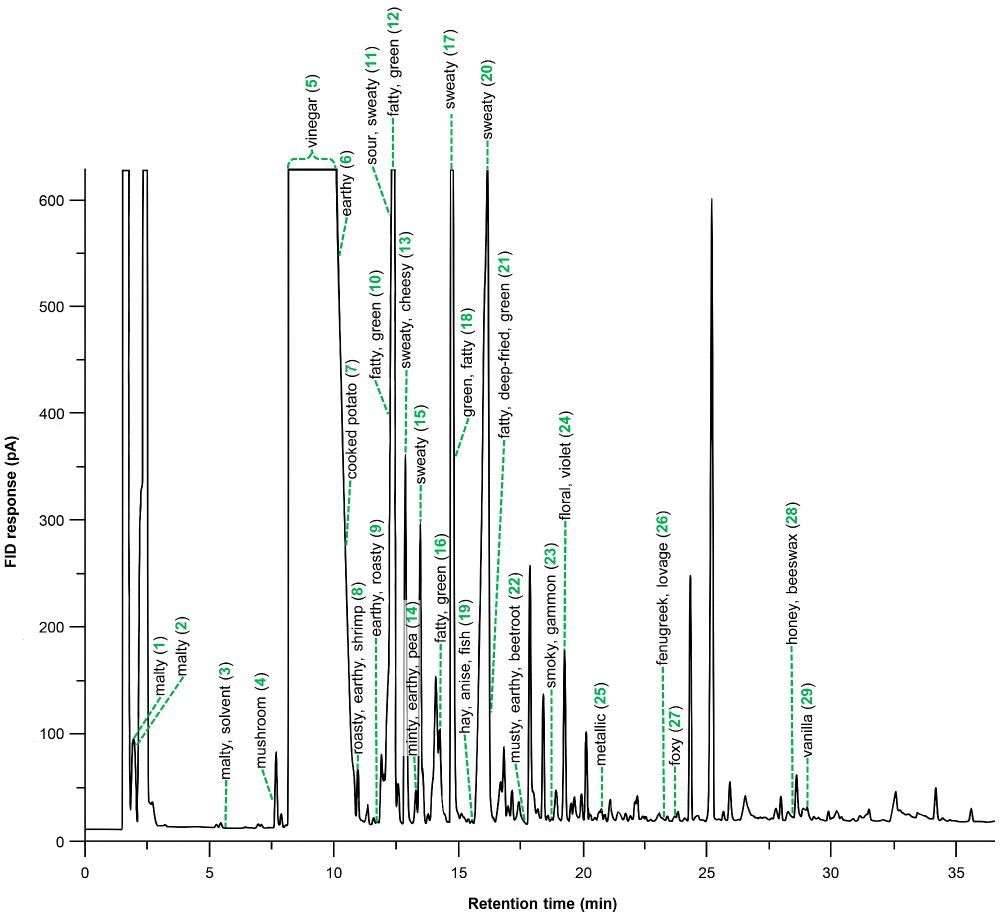
Major highlight: 85% Synechococcus proteome coverage in a single shotgun experiment.
Out now in Plant Physiology!
doi.org/10.1093/plph...

Major highlight: 85% Synechococcus proteome coverage in a single shotgun experiment.
Out now in Plant Physiology!
doi.org/10.1093/plph...
thank you to all the authors!
www.pnas.org/doi/10.1073/...

thank you to all the authors!
www.pnas.org/doi/10.1073/...
doi.org/10.3390/ijms...
doi.org/10.3390/ijms...
Find it here🔽
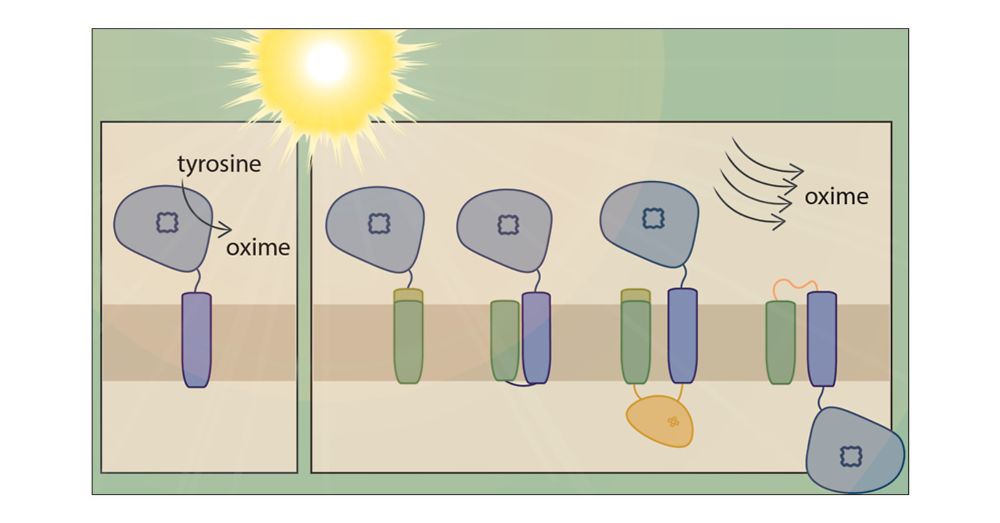
Great collab with @synbiojazz.bsky.social!
#teamgreen #cyanobacteria
academic.oup.com/sumbio/artic...

Great collab with @synbiojazz.bsky.social!
#teamgreen #cyanobacteria
academic.oup.com/sumbio/artic...


Towards the control of biofilm formation in Anabaena (Nostoc) sp. PCC7120: novel insights into the genes involved and their regulation. nph.onlinelibrary.wiley.com/doi/10.1111/...

Towards the control of biofilm formation in Anabaena (Nostoc) sp. PCC7120: novel insights into the genes involved and their regulation. nph.onlinelibrary.wiley.com/doi/10.1111/...
👉 buff.ly/jWqcD6B \
#PlantScience

👉 buff.ly/jWqcD6B \
#PlantScience
Warm congratulations @lauratwey.bsky.social! 🌷
www.utu.fi/en/news/pres...




www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
We have been thinking about this for a long time (nanoLC, capLC, µLC). According to a new @biorxivpreprint.bsky.social by the @kusterlab.bsky.social, it seems like all options work very well. www.biorxiv.org/cont...

We have been thinking about this for a long time (nanoLC, capLC, µLC). According to a new @biorxivpreprint.bsky.social by the @kusterlab.bsky.social, it seems like all options work very well. www.biorxiv.org/cont...

Excited to share our latest study, where we uncovered a previously unknown bacterial defense system in M. #tuberculosis: effluxosomes — dynamic membrane clusters that coordinate resistance to multiple toxic metals.
shorturl.at/Ek0HD
#MicroSky
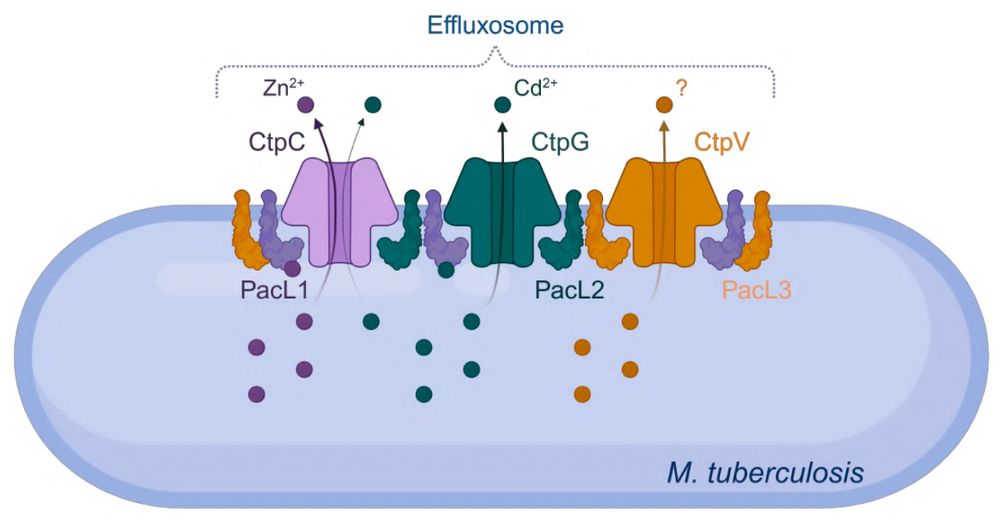
Excited to share our latest study, where we uncovered a previously unknown bacterial defense system in M. #tuberculosis: effluxosomes — dynamic membrane clusters that coordinate resistance to multiple toxic metals.
shorturl.at/Ek0HD
#MicroSky
pubs.acs.org/doi/10.1021/...