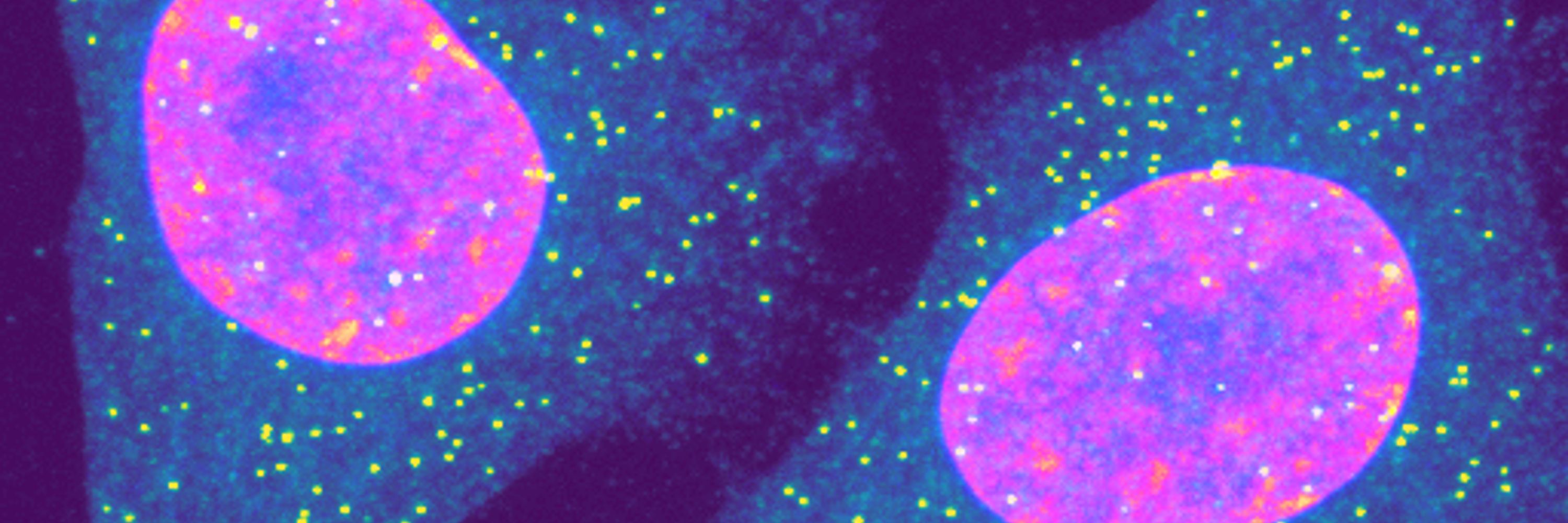
Studying transcription factors & promoters
University of Milan 🇮🇹
previously in Tora's lab, IGBMC 🇫🇷
Q-rich activation domains: flexible ‘rulers’ for transcription start site selection?
www.cell.com/trends/genet...

(TLDR; low-affinity motifs matter as pioneers!)

(TLDR; low-affinity motifs matter as pioneers!)
doi.org/10.1016/j.ce...
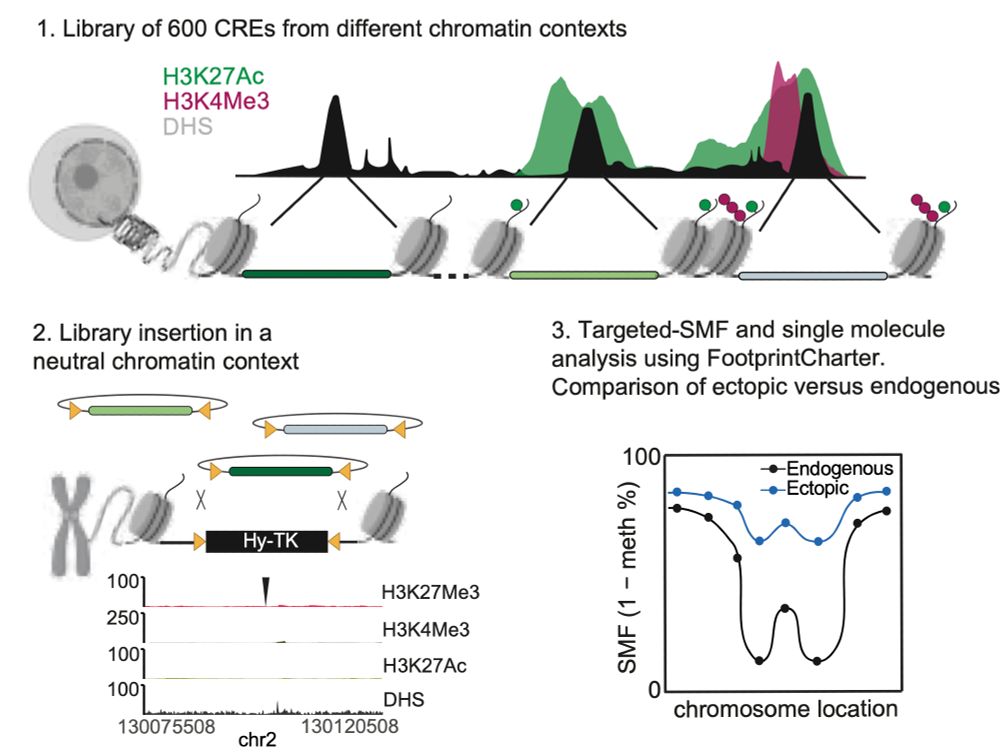
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
Our new preprint reveals how divergence in TBP’s domains shapes transcriptional specialization across eukaryotes.
Read it here 👉
www.biorxiv.org/content/10.1...
#MolecularEvolution #Transcription

Our new preprint reveals how divergence in TBP’s domains shapes transcriptional specialization across eukaryotes.
Read it here 👉
www.biorxiv.org/content/10.1...
#MolecularEvolution #Transcription
Cell-type-specific functionality encoded within the intrinsically disordered regions of OCT4
www.nature.com/articles/s41...

Cell-type-specific functionality encoded within the intrinsically disordered regions of OCT4
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...



www.biorxiv.org/content/10.1... 1/n

www.biorxiv.org/content/10.1... 1/n
www.cell.com/molecular-ce...
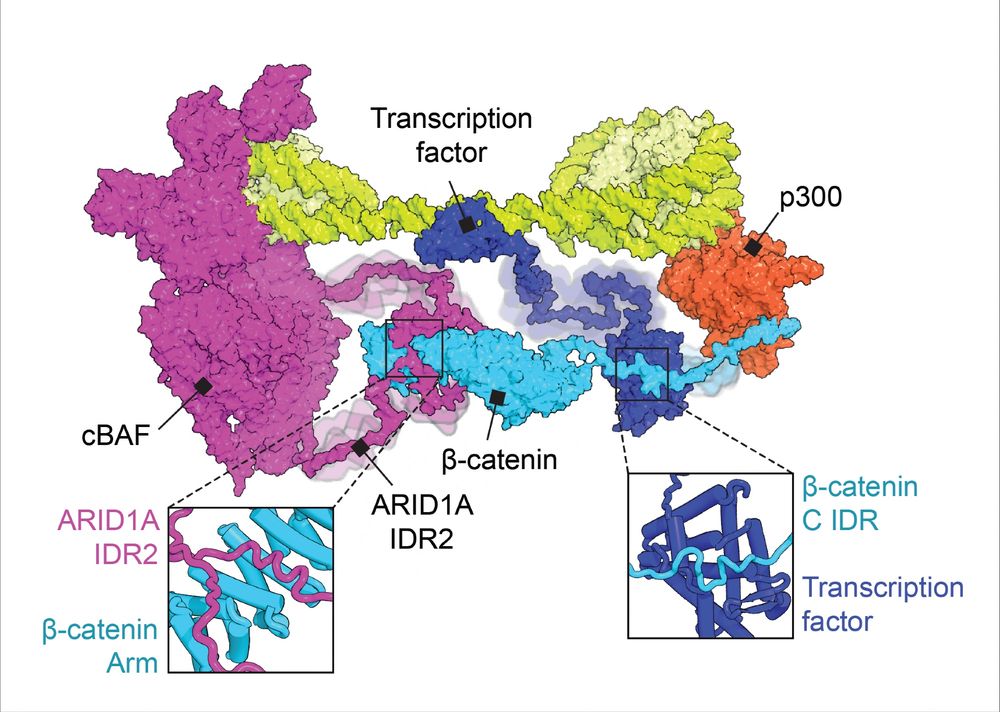
www.cell.com/molecular-ce...
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
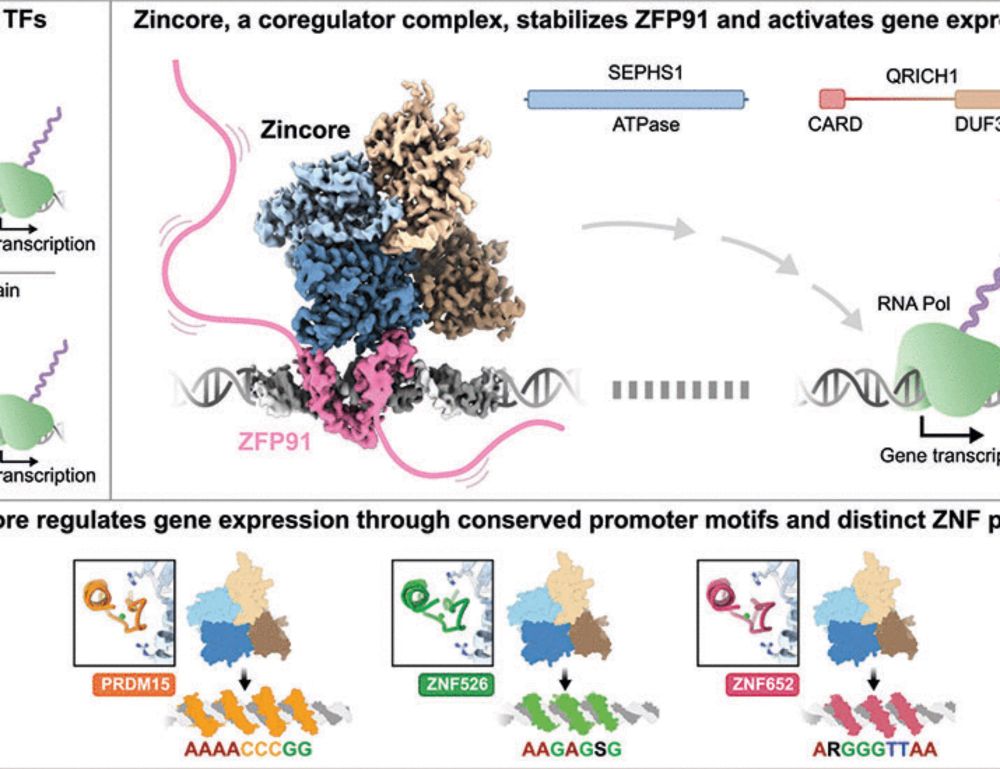
🔗 doi.org/10.1093/gbe/evaf071
#genome #evolution #transcription #TEsky

🔗 doi.org/10.1093/gbe/evaf071
#genome #evolution #transcription #TEsky
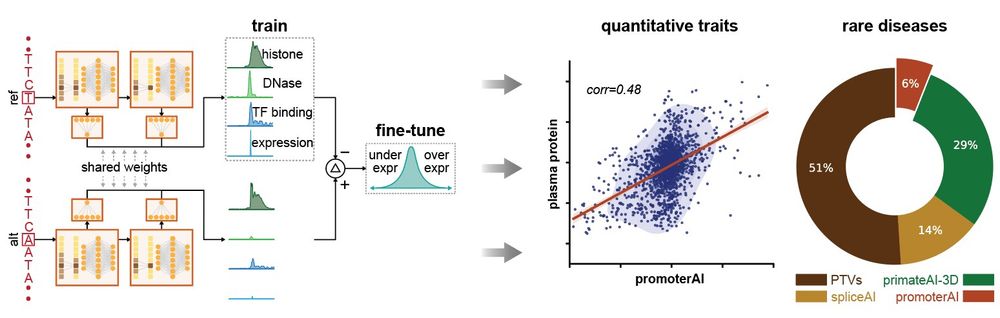
New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics

New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics
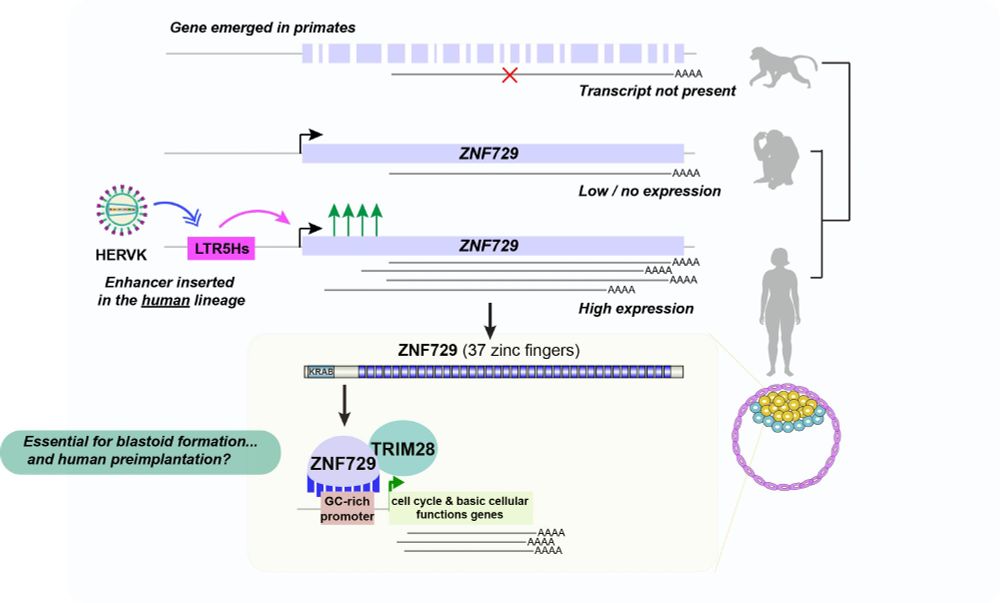
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

The first #CryoEM structure of a Mediator subunit (MED23) in complex with the transactivation domain of Elk-1. Unstructured TAD of course, and to spice things up, it's phosphorylation dependent.
www.nature.com/articles/s41...

The first #CryoEM structure of a Mediator subunit (MED23) in complex with the transactivation domain of Elk-1. Unstructured TAD of course, and to spice things up, it's phosphorylation dependent.
www.nature.com/articles/s41...
Topics include genomic language models, hyperdivergent regions, meiotic DSBs, cfDNA in pop’n genetics, genital evolution, ‘rulers’ for TSS selection, spatial multiomics in embryos, and more:
www.cell.com/trends/genet...
Thanks to @yun-s-song.bsky.social for the cover!

Topics include genomic language models, hyperdivergent regions, meiotic DSBs, cfDNA in pop’n genetics, genital evolution, ‘rulers’ for TSS selection, spatial multiomics in embryos, and more:
www.cell.com/trends/genet...
Thanks to @yun-s-song.bsky.social for the cover!
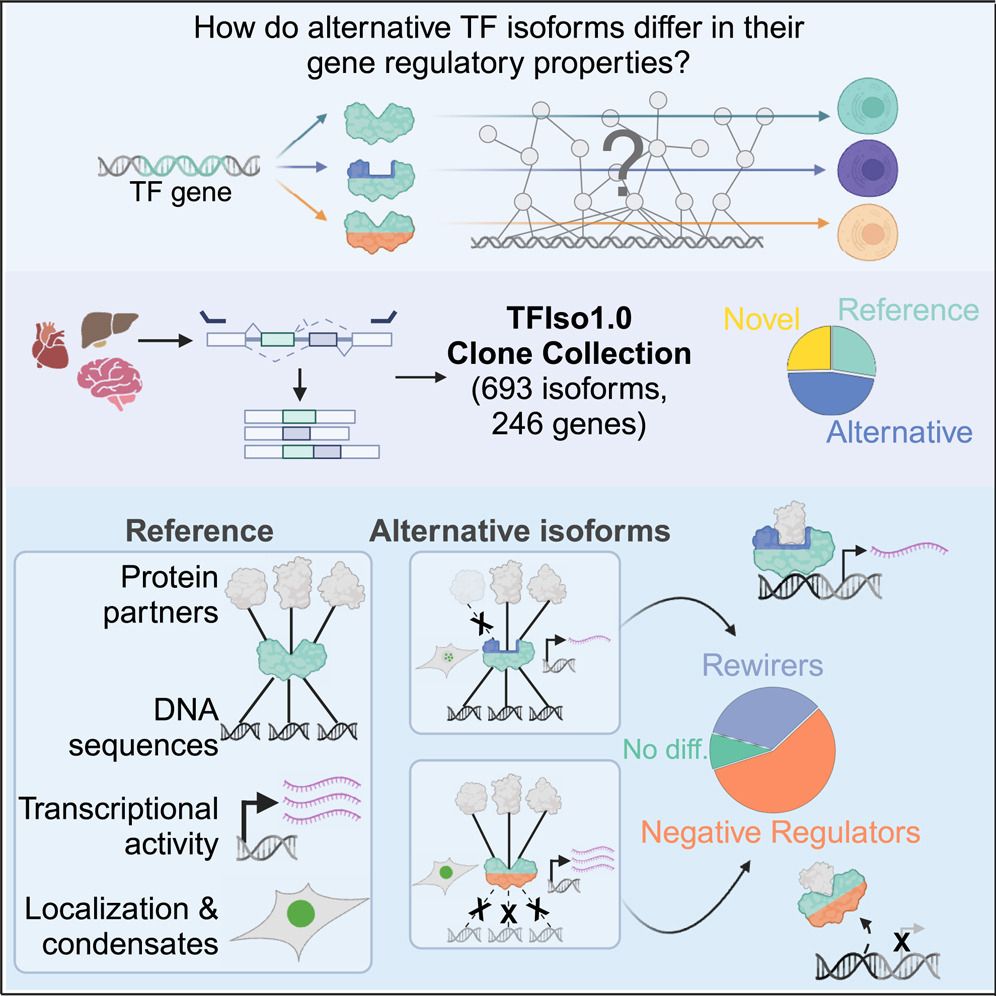
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...

