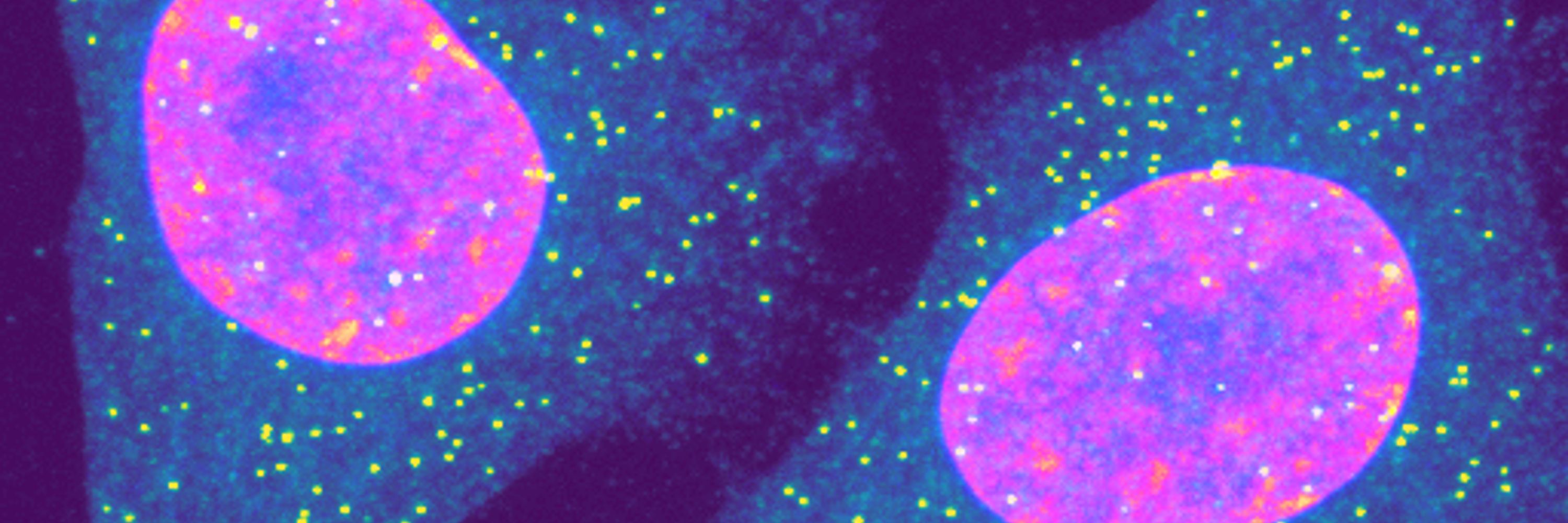
Studying transcription factors & promoters
University of Milan 🇮🇹
previously in Tora's lab, IGBMC 🇫🇷





doi.org/10.1016/j.jm...
1/5

doi.org/10.1016/j.jm...
1/5
doi.org/10.1016/j.ti...

doi.org/10.1016/j.ti...



















