@MichiganEarth and graft vs. host disease @umichmedicine
PhD @UDelaware, 🦠 in/on concrete
doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6

doi.org/10.1038/s415...
@benjwoodcroft.bsky.social @rhysnewell.bsky.social
🧵1/6
Plasmidsaurus now offers RNA sequencing for gene expression analysis:
• As fast as 3 day turnaround
• $50/sample for academia, $80 for industry
• Up to ~10M unique transcript 3’ end reads per sample
• Interactive results
Explore Plasmidsaurus RNA-Seq today.

Plasmidsaurus now offers RNA sequencing for gene expression analysis:
• As fast as 3 day turnaround
• $50/sample for academia, $80 for industry
• Up to ~10M unique transcript 3’ end reads per sample
• Interactive results
Explore Plasmidsaurus RNA-Seq today.

Check out their manuscript here:
www.nature.com/articles/s41...

Check out their manuscript here:
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...

@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
- 30-40% potential reduction in memory with approximately the same runtime.
- Breaking changes to indexing and searching databases
Calculate ANI for contigs, genomes -- even search > 140k genomes. Pre-indexed GTDB-R226 available for download.

- 30-40% potential reduction in memory with approximately the same runtime.
- Breaking changes to indexing and searching databases
Calculate ANI for contigs, genomes -- even search > 140k genomes. Pre-indexed GTDB-R226 available for download.
pubs.acs.org/doi/abs/10.1...
pubs.acs.org/doi/abs/10.1...
myloasm-docs.github.io
myloasm-docs.github.io
By @annaleighclark.bsky.social

By @annaleighclark.bsky.social
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

GTDB - Genome Taxonomy Database https://gtdb.ecogenomic.org/
GTDB - Genome Taxonomy Database https://gtdb.ecogenomic.org/
2. The packages don’t exist. Which would normally cause an error but
3. Nefarious people have made malware under the package names that LLMs make up most often. So
4. Now the LLM code points to malware.

2. The packages don’t exist. Which would normally cause an error but
3. Nefarious people have made malware under the package names that LLMs make up most often. So
4. Now the LLM code points to malware.
Its near-final budget proposal would end all NOAA research labs, academic institutes, and regional climate centers. And it wants to fully end the NOAA Research division.

Its near-final budget proposal would end all NOAA research labs, academic institutes, and regional climate centers. And it wants to fully end the NOAA Research division.
ES&T
"Genomic Identification and Characterization of Saxitoxin Producing Cyanobacteria in Western Lake Erie Harmful Algal Blooms" doi.org/10.1021/acs....
@GreatLakesGreg @pubs.acs.org #greatlakes #ciglr #noaaglerl

ES&T
"Genomic Identification and Characterization of Saxitoxin Producing Cyanobacteria in Western Lake Erie Harmful Algal Blooms" doi.org/10.1021/acs....
@GreatLakesGreg @pubs.acs.org #greatlakes #ciglr #noaaglerl
Retroactive to February 🤡
Retroactive to February 🤡
ⓁⓄⒸⒶⓉⒾⓄⓃ
ⓁⓄⒸⒶⓉⒾⓄⓃ
ⓁⓄⒸⒶⓉⒾⓄⓃ
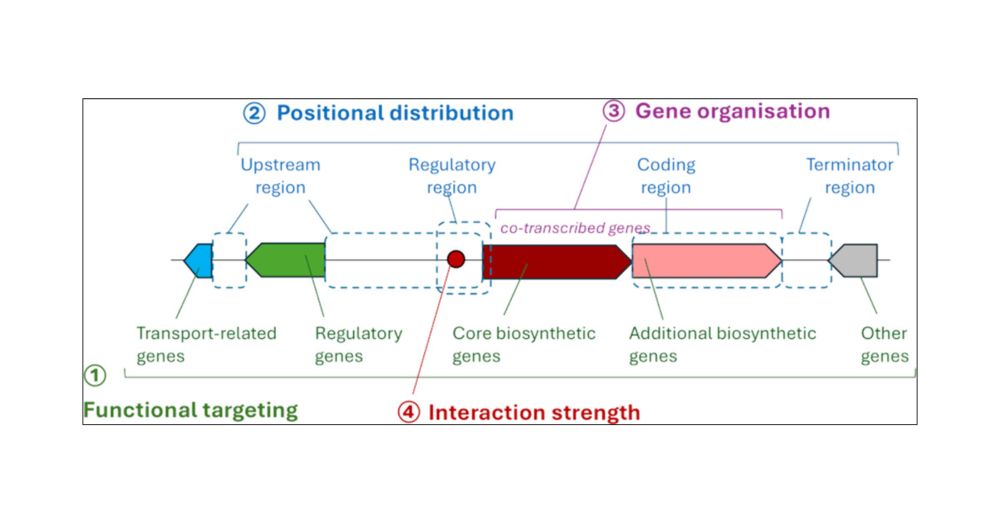
Thought bacterial gene regulation is all about promoters and repressors?
𝙏𝙃𝙄𝙉𝙆 𝘼𝙂𝘼𝙄𝙉
WHERE a gene sits on the chromosome can dictate its expression—especially in fast growing bacteria
Evolution won’t leave genome layout to chance
Hu et al SCIENCE

ⓁⓄⒸⒶⓉⒾⓄⓃ
ⓁⓄⒸⒶⓉⒾⓄⓃ
ⓁⓄⒸⒶⓉⒾⓄⓃ
Thought bacterial gene regulation is all about promoters and repressors?
𝙏𝙃𝙄𝙉𝙆 𝘼𝙂𝘼𝙄𝙉
WHERE a gene sits on the chromosome can dictate its expression—especially in fast growing bacteria
Evolution won’t leave genome layout to chance
Hu et al SCIENCE
CoverM: Read alignment statistics for metagenomics academic.oup.com/bioinformati... #jcampubs

CoverM: Read alignment statistics for metagenomics academic.oup.com/bioinformati... #jcampubs


