
Microbial, viral, & geochemical interactions by linking meta'omics & microscopy
Managed by @geomicrosoares.bsky.social
@geomicrosoares.bsky.social @feriel.bsky.social @pengyaoz.bsky.social are out in Regensburg!
@unidue.bsky.social @uniregensburg.bsky.social


@geomicrosoares.bsky.social @feriel.bsky.social @pengyaoz.bsky.social are out in Regensburg!
@unidue.bsky.social @uniregensburg.bsky.social
This provides an explanation for some of the many unmatched spacers in public databases.
@unidue.bsky.social
This provides an explanation for some of the many unmatched spacers in public databases.
@unidue.bsky.social
Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!

Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!
@unidue.bsky.social

@unidue.bsky.social
Check out their manuscript here:
www.nature.com/articles/s41...

Check out their manuscript here:
www.nature.com/articles/s41...
Shoutout to the remainder collaborators at the China University of Geosciences, and Jon Lloyd at the University of Manchester (@manchesterup.bsky.social).
@unidue.bsky.social & others RT!
Shoutout to the remainder collaborators at the China University of Geosciences, and Jon Lloyd at the University of Manchester (@manchesterup.bsky.social).
@unidue.bsky.social & others RT!
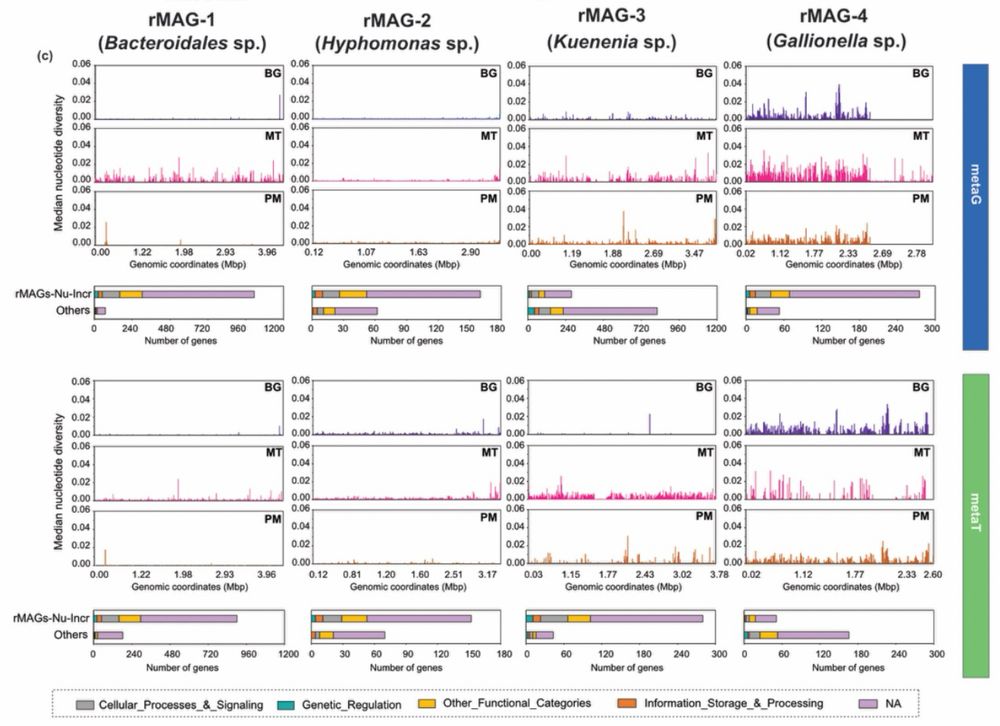
We show that U in-situ leaching drives significant alterations in 🦠 community composition, oxidative stress, biogeochemical cycling, and overexpression of 🧬 repair genes

We show that U in-situ leaching drives significant alterations in 🦠 community composition, oxidative stress, biogeochemical cycling, and overexpression of 🧬 repair genes
We're happy to see this preprint out, fruit of a recent collaboration with @wei-biogeochem.bsky.social! 🧬💻
Small 🧵 below going through the main points 👇
www.biorxiv.org/content/10.1...

We're happy to see this preprint out, fruit of a recent collaboration with @wei-biogeochem.bsky.social! 🧬💻
Small 🧵 below going through the main points 👇
www.biorxiv.org/content/10.1...
This PhD will take place in close collaboration with the @jgi.doe.gov. 🦠🧬💻
Apply here: www.uni-due.de/karriere/ste...

Finally, thank you @leeselab.bsky.social for sitting in the thesis committee! 🫶

Finally, thank you @leeselab.bsky.social for sitting in the thesis committee! 🫶

Read more below:
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...
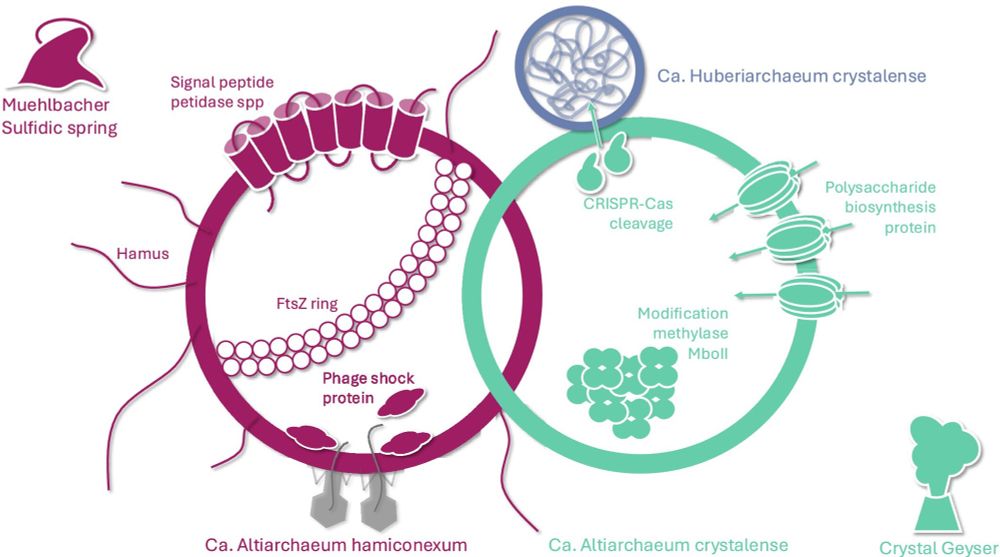
Read more below:
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...

@crc-resist.bsky.social @unidue.bsky.social


@crc-resist.bsky.social @unidue.bsky.social

On our side, we’re exceptionally giving two talks about our latest work:
⬇️
On our side, we’re exceptionally giving two talks about our latest work:
⬇️

@unidue.bsky.social
@unidue.bsky.social
See our latest publication: doi.org/10.1093/isme...

See our latest publication: doi.org/10.1093/isme...
With @TheWrightonLab at CSU, we built river MAG & gene DBs and used metaT data to explore this while uncovering active stream microbiome metabolic functions!
Read our preprint here: doi.org/10.1101/2025...
@unidue.bsky.social

With @TheWrightonLab at CSU, we built river MAG & gene DBs and used metaT data to explore this while uncovering active stream microbiome metabolic functions!
Read our preprint here: doi.org/10.1101/2025...
@unidue.bsky.social

