
Happy to share our recent publication: Quantitative modeling of mRNA degradation reveals tempo-dependent mRNA clearance in early embryos
(1/7)
academic.oup.com/nar/article/...

Happy to share our recent publication: Quantitative modeling of mRNA degradation reveals tempo-dependent mRNA clearance in early embryos
(1/7)
academic.oup.com/nar/article/...
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
So incredibly proud of @tobiaswilliams.bsky.social & Ewa Michalak who led the work!
www.cell.com/molecular-ce...
So incredibly proud of @tobiaswilliams.bsky.social & Ewa Michalak who led the work!
www.cell.com/molecular-ce...
tinyurl.com/setd2
tinyurl.com/setd2
Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻

Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
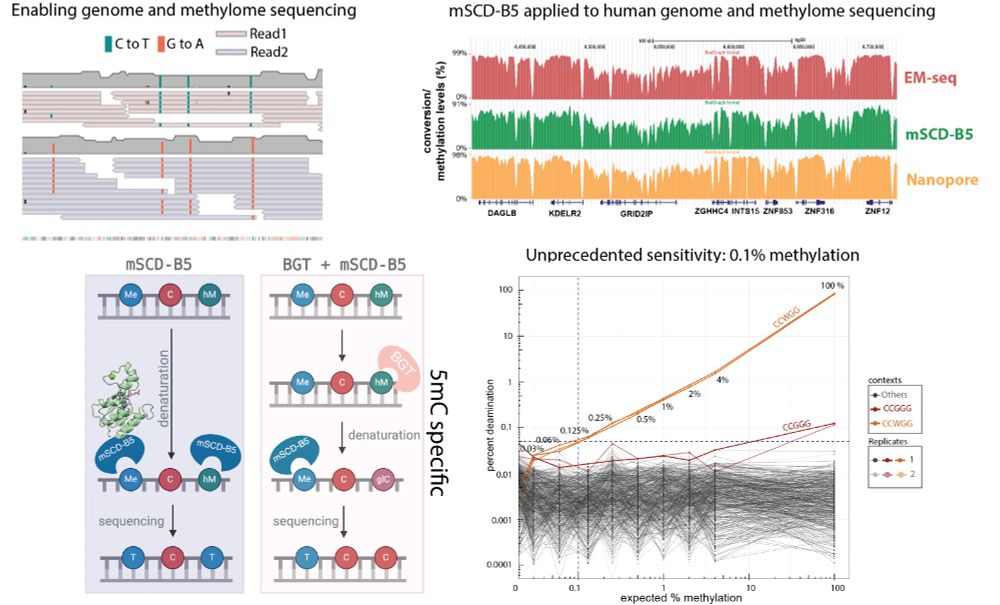
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
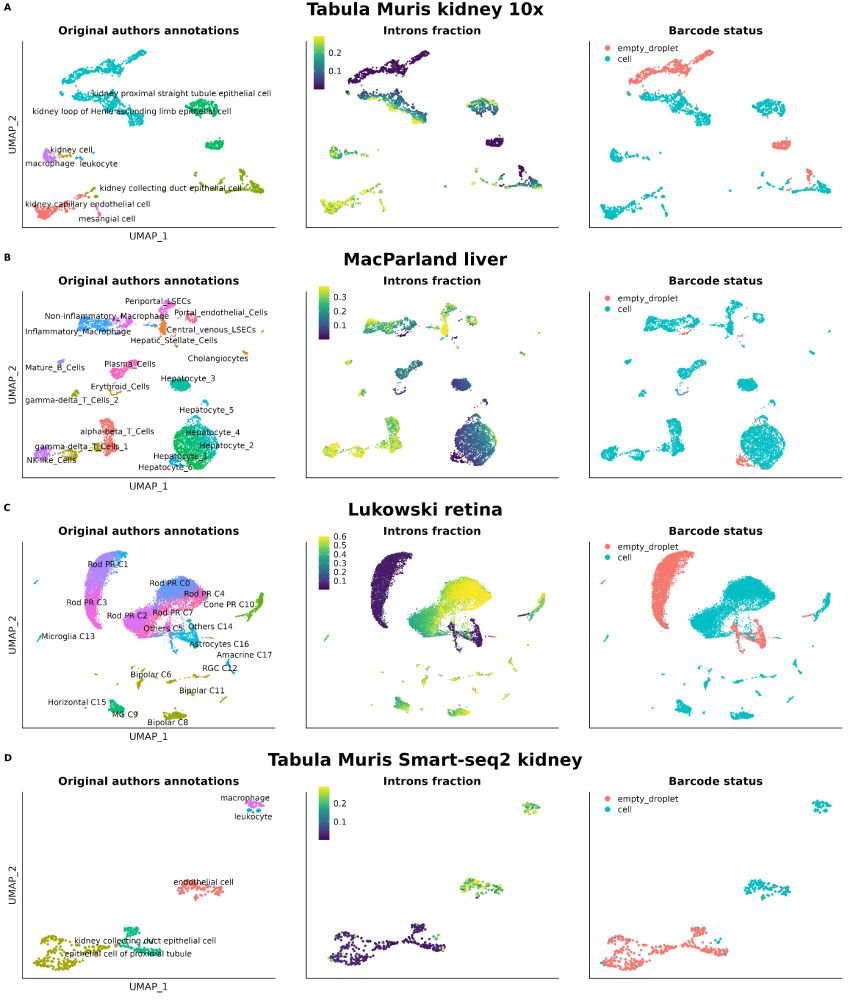
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
Why? And why now?
direct.mit.edu/qss/article/...
1/n 🧵

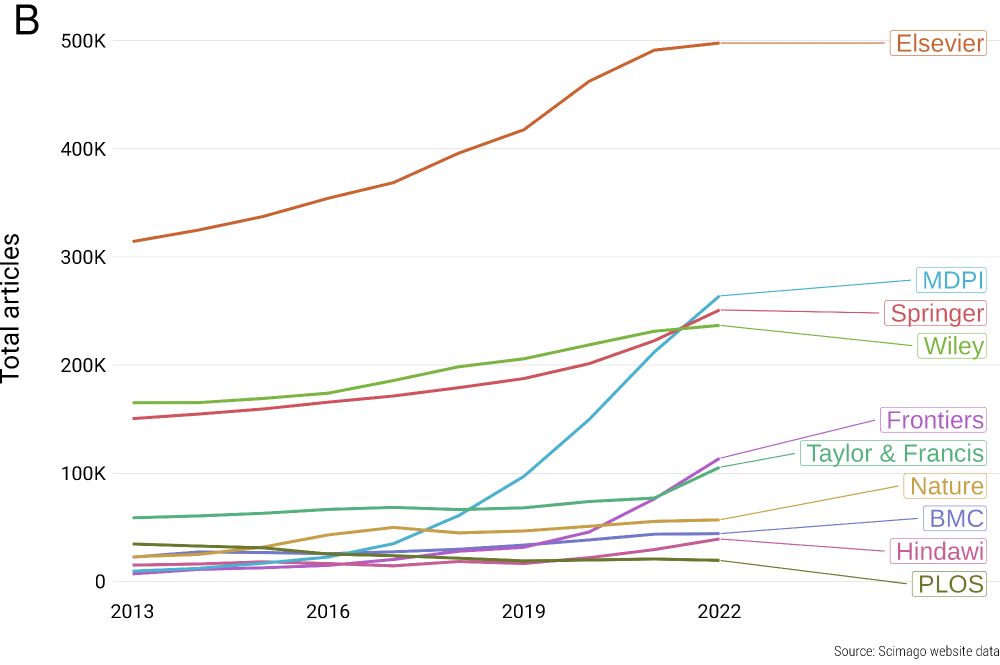
Why? And why now?
direct.mit.edu/qss/article/...
1/n 🧵
🏔️ logan-search.org 🏔️
#Genomics #Bioinformatics #OpenScience

🏔️ logan-search.org 🏔️
#Genomics #Bioinformatics #OpenScience
www.cell.com/molecular-ce...

www.cell.com/molecular-ce...
#chromatin #enhancers
www.cell.com/molecular-ce...

#chromatin #enhancers
www.cell.com/molecular-ce...
www.nature.com/articles/s41...
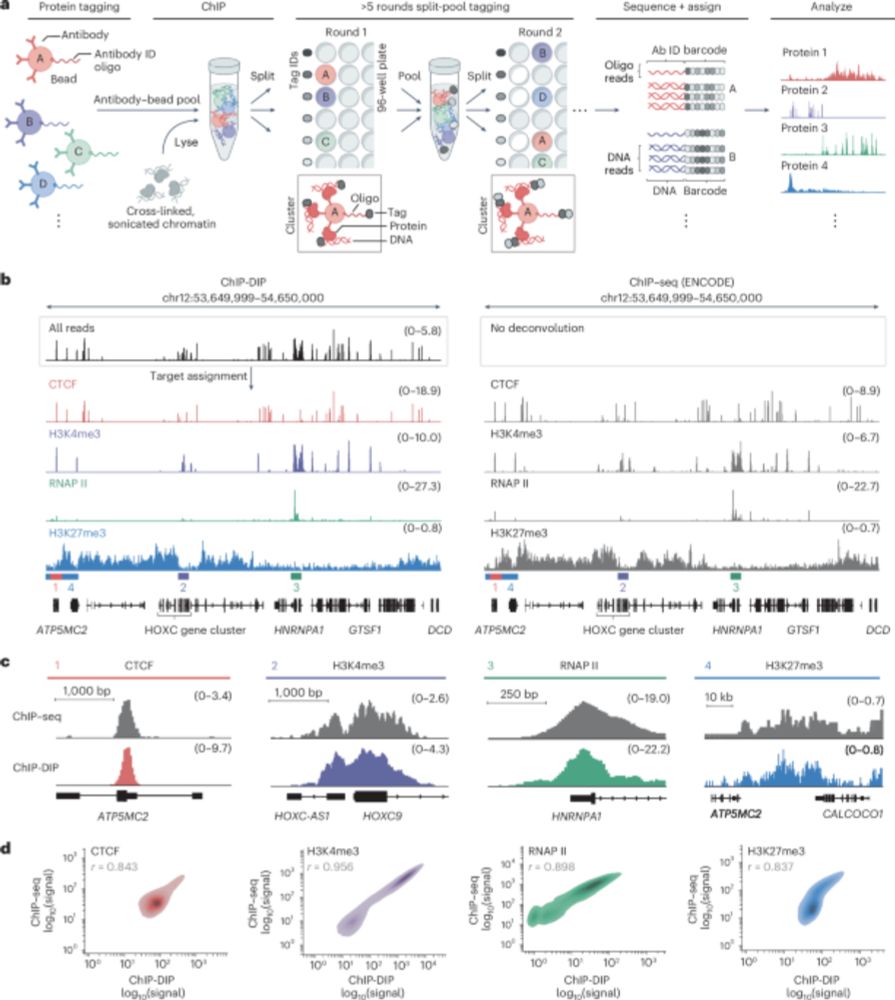
www.nature.com/articles/s41...
go.bsky.app/QVPoZXp
go.bsky.app/QVPoZXp
www.nature.com/articles/s43...

www.nature.com/articles/s43...
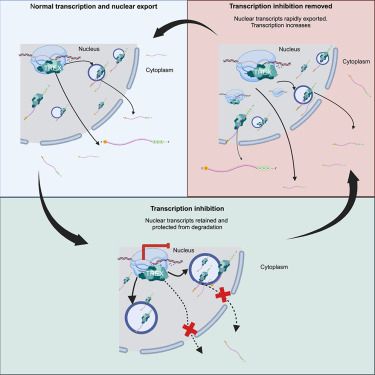
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
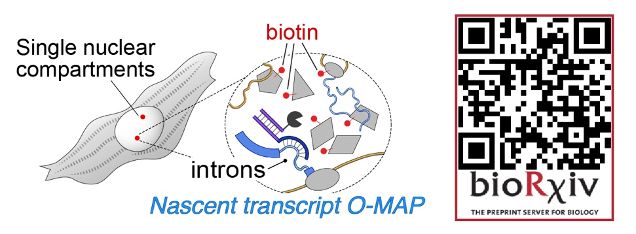
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)

Inferring DNA methylation in non-skeletal tissues of ancient specimens. A major challenge in studying DNA methylation in ancient samples is that it is tissue-specific.
Inferring DNA methylation in non-skeletal tissues of ancient specimens. A major challenge in studying DNA methylation in ancient samples is that it is tissue-specific.
www.cell.com/action/showP...
www.cell.com/action/showP...
Available on bioconductor: t.co/CyK0YVokV9 and github: t.co/EzSWAUFeel
Available on bioconductor: t.co/CyK0YVokV9 and github: t.co/EzSWAUFeel

www.embopress.org/doi/full/10....
#epigenetics

www.embopress.org/doi/full/10....
#epigenetics


