
Haixin Wang and Shengquan Chen reporting in Scientific Data
#Epigenetics #DNAm #DNAme #Aging
---
Empower your research with high-res epigenetic insights at epigenometech.com

Haixin Wang and Shengquan Chen reporting in Scientific Data
#Epigenetics #DNAm #DNAme #Aging
---
Empower your research with high-res epigenetic insights at epigenometech.com
www.biorxiv.org/content/10.1...
🧵 below

www.biorxiv.org/content/10.1...
🧵 below
www.nature.com/articles/s41...
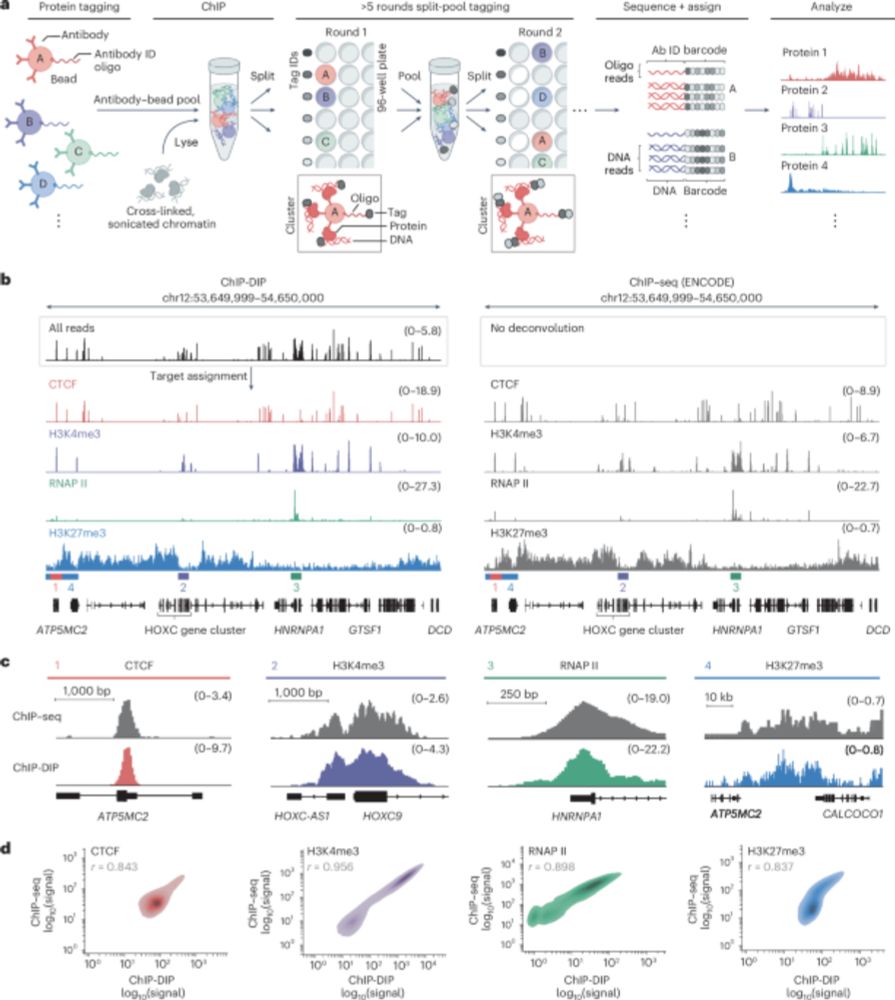
www.nature.com/articles/s41...
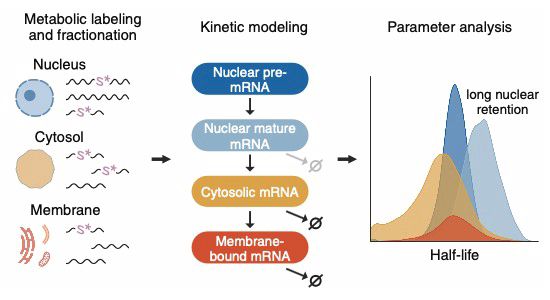
www.nature.com/articles/s41...
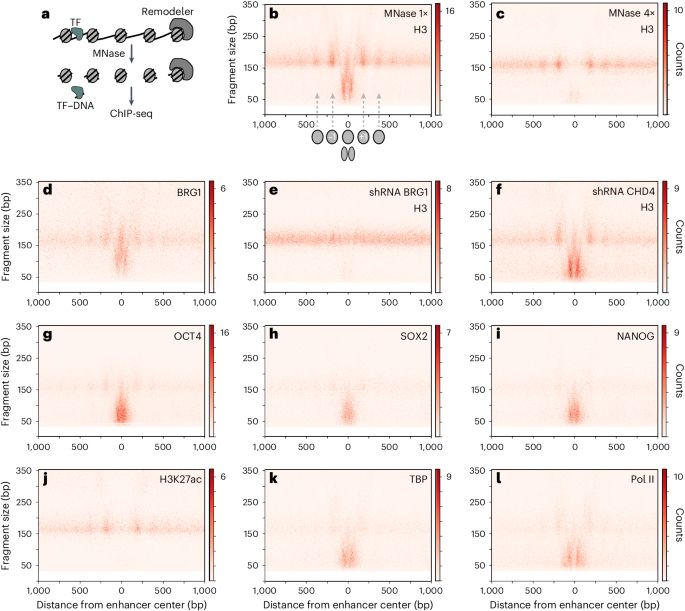
www.nature.com/articles/s41...
Link: doi.org/10.1016/j.cmet.2024.06.006
Thread 🧵👇1/9

Link: doi.org/10.1016/j.cmet.2024.06.006
Thread 🧵👇1/9
"How Margaret Dayhoff Brought Modern Computing to Biology"
www.smithsonianmag.com/science-natu...
"How Margaret Dayhoff Brought Modern Computing to Biology"
www.smithsonianmag.com/science-natu...
www.cell.com/action/showP...
www.cell.com/action/showP...
New preprint from a collaboration led by Grace Bower and Evgeny Kvon.
doi.org/10.1101/2024...
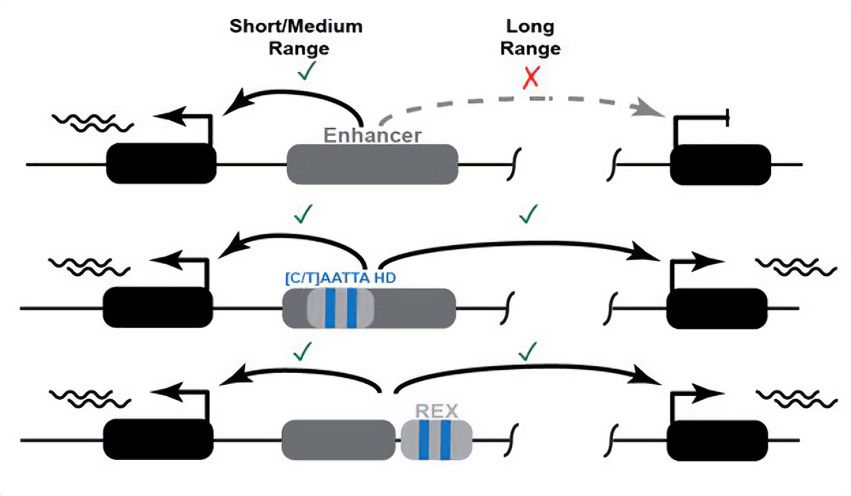
www.nature.com/articles/s41...

www.nature.com/articles/s41...
go.bsky.app/StHb8Ma
go.bsky.app/StHb8Ma
go.bsky.app/9jdDNxG
go.bsky.app/9jdDNxG
Let me know if you would like to be added.
go.bsky.app/6tTQdqQ
Let me know if you would like to be added.
go.bsky.app/6tTQdqQ

