https://lotharukpongjs.github.io/
New paper from my postdoc @mpi-bio-fml.bsky.social out in PNAS:
Germline fate determination by a single ARGONAUTE protein in Ectocarpus www.pnas.org/doi/10.1073/...

New paper from my postdoc @mpi-bio-fml.bsky.social out in PNAS:
Germline fate determination by a single ARGONAUTE protein in Ectocarpus www.pnas.org/doi/10.1073/...
🎓 www.cell.com/cell/fulltex...
Plants and animals evolve radically different body plans.
Do they also operate under fundamentally different molecular evolutionary constraints during organ formation?
@cellpress.bsky.social

🎓 www.cell.com/cell/fulltex...
Plants and animals evolve radically different body plans.
Do they also operate under fundamentally different molecular evolutionary constraints during organ formation?
@cellpress.bsky.social
Excited to share this highly efficient, transgene-free CRISPR–Cas genome editing protocol for brown algae, requiring no cloning and no specialized equipment.
doi.org/10.1016/j.cr...
#CRISPR #BrownAlgae
@mpi-bio-fml.bsky.social

Excited to share this highly efficient, transgene-free CRISPR–Cas genome editing protocol for brown algae, requiring no cloning and no specialized equipment.
doi.org/10.1016/j.cr...
#CRISPR #BrownAlgae
@mpi-bio-fml.bsky.social

🌱Check out our new Opinion Paper with @thanvisrikant.bsky.social discussing this topic in @cp-trendsplantsci.bsky.social!
www.sciencedirect.com/science/arti...

🌱Check out our new Opinion Paper with @thanvisrikant.bsky.social discussing this topic in @cp-trendsplantsci.bsky.social!
www.sciencedirect.com/science/arti...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

go.nature.com/4i0i61w

Is it sponges (panels A & B) or comb jellies (C & D) that root the animal tree of life?
For over 15 years, #phylogenomic studies have been divided.
We provide new evidence suggesting that...
🔗: www.science.org/doi/10.1126/...

Is it sponges (panels A & B) or comb jellies (C & D) that root the animal tree of life?
For over 15 years, #phylogenomic studies have been divided.
We provide new evidence suggesting that...
🔗: www.science.org/doi/10.1126/...
Now with long-awaited parallel distance computation & a full speed-optimized refactor thanks to Andrew Gene Brown.
Compute 50+ distances/divergences in R faster than ever.
📦 CRAN: cran.r-project.org/web/packages...
💻 Code: github.com/drostlab/phi...

Now with long-awaited parallel distance computation & a full speed-optimized refactor thanks to Andrew Gene Brown.
Compute 50+ distances/divergences in R faster than ever.
📦 CRAN: cran.r-project.org/web/packages...
💻 Code: github.com/drostlab/phi...
genzplyr is dplyr, but bussin fr fr no cap.

genzplyr is dplyr, but bussin fr fr no cap.
www.nature.com/articles/s41...

www.nature.com/articles/s41...
genomebiology.biomedcentral.com/articles/10....
A wonderful collaboration with @ericadinatale.bsky.social, @cssmartinho.bsky.social, @rorycraig.bsky.social, and Susana Coelho! 🎉

genomebiology.biomedcentral.com/articles/10....
A wonderful collaboration with @ericadinatale.bsky.social, @cssmartinho.bsky.social, @rorycraig.bsky.social, and Susana Coelho! 🎉
A short thread on how Ectocarpus and its TE secrets have kept me busy lately:
rdcu.be/eITQH
A short thread on how Ectocarpus and its TE secrets have kept me busy lately:
rdcu.be/eITQH
doi.org/10.1101/2025...
A thread 👇

doi.org/10.1101/2025...
A thread 👇
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
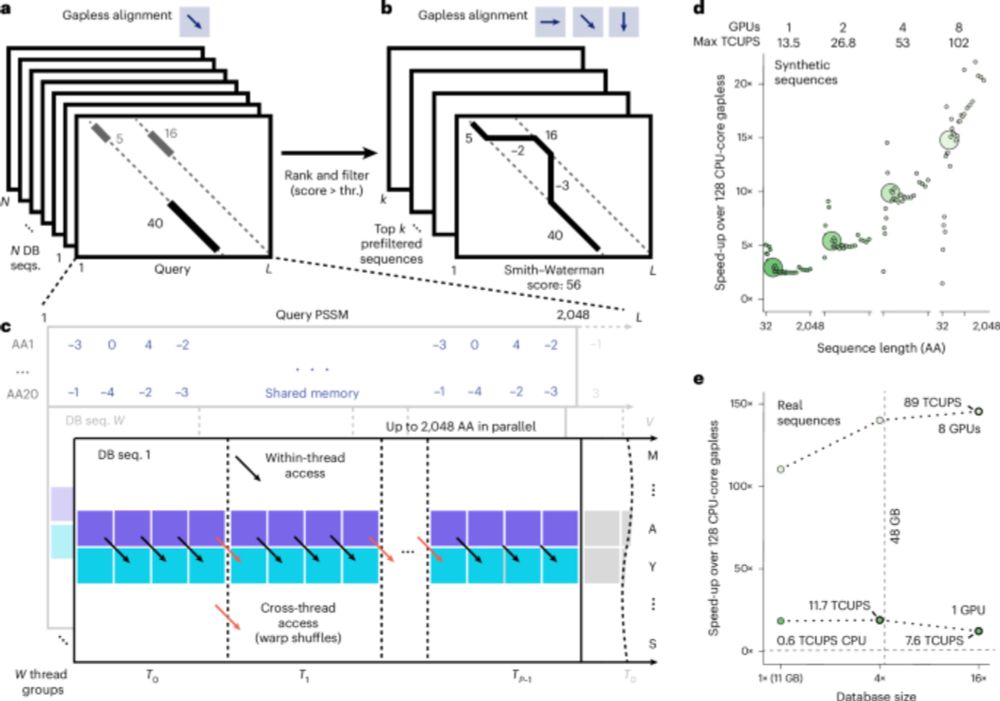
www.nature.com/articles/s41...
It's not every day we have a new major #ggplot2 release but it is a fitting 18 year birthday present for the package.
Get an overview of the release in this blog post and be on the lookout for more in-depth posts #rstats

In our paper out now in @currentbiology.bsky.social we show that the Atlas blue butterfly has 229 chromosome pairs- the highest in diploid Metazoa! These arose by rapid autosome fragmentation while sex chromosomes stayed intact.
www.cell.com/current-biol...
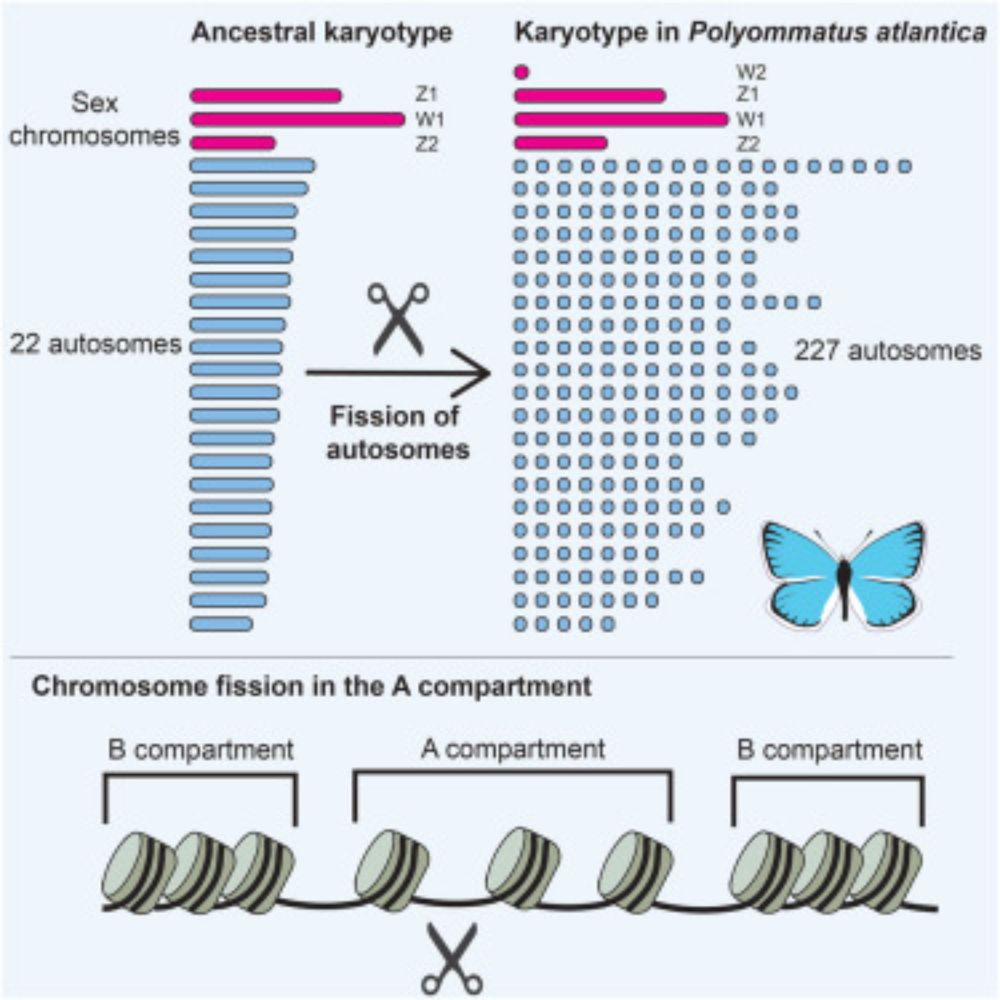
In our paper out now in @currentbiology.bsky.social we show that the Atlas blue butterfly has 229 chromosome pairs- the highest in diploid Metazoa! These arose by rapid autosome fragmentation while sex chromosomes stayed intact.
www.cell.com/current-biol...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
doi.org/10.1186/s130...

doi.org/10.1186/s130...
www.nature.com/articles/s41...

with #LauraPiovani @dariagavr.bsky.social @alexdemendoza.bsky.social @chemamd.bsky.social and others /1

with #LauraPiovani @dariagavr.bsky.social @alexdemendoza.bsky.social @chemamd.bsky.social and others /1
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
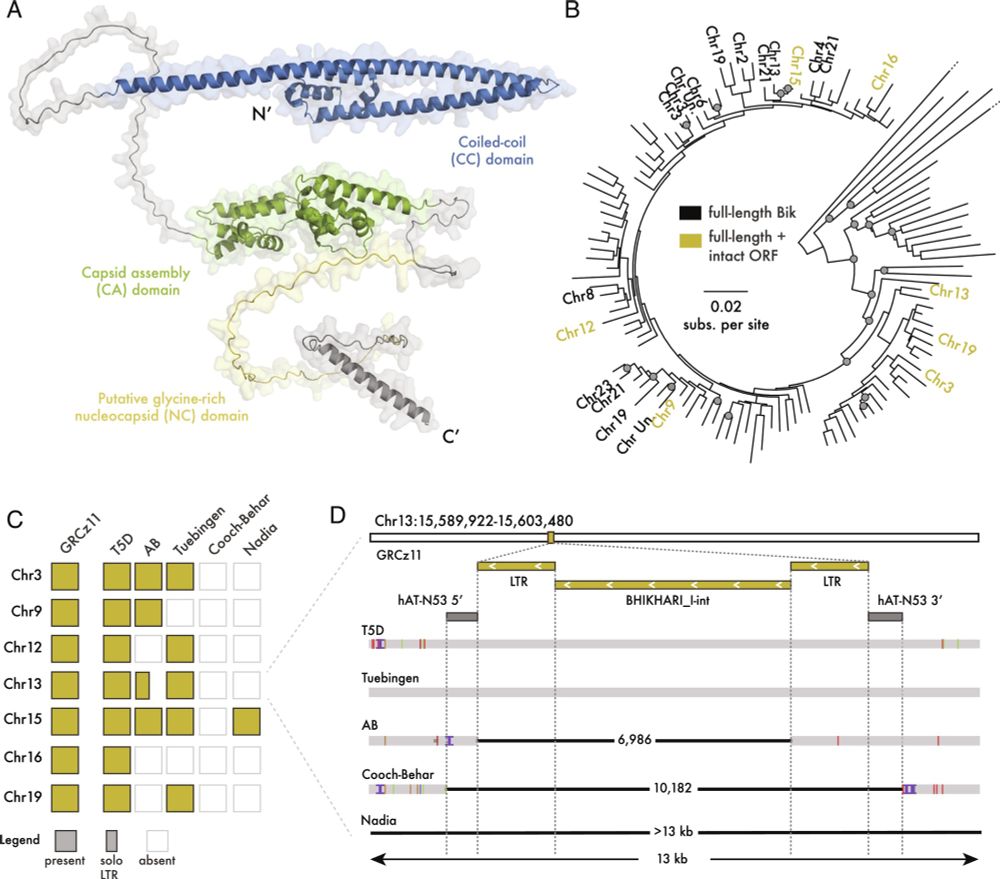
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n

