RECOMB-CG: 13 February
RECOMB-RSG: 15 February
RECOMB-Privacy: 9 March
RECOMB-Seq: 12 March (abstract registration)
RECOMB-Arch: 12 March (abstract registration)
RECOMB-Genetics: 13 March
#RECOMB2026 #deadlines
RECOMB-CG: 13 February
RECOMB-RSG: 15 February
RECOMB-Privacy: 9 March
RECOMB-Seq: 12 March (abstract registration)
RECOMB-Arch: 12 March (abstract registration)
RECOMB-Genetics: 13 March
#RECOMB2026 #deadlines

I heard you craving for more combinatorics, here are some more for y'all !
I heard you craving for more combinatorics, here are some more for y'all !
arxiv.org/abs/2602.03525
TLDR:
ZOR filters are STATIC filters with false positives.
-Almost memory optimal: <1% overhead over the theoretical lower bound (!!!)
-Fast queries: ~100 ns
-Construction cannot fail
A thread:

arxiv.org/abs/2602.03525
TLDR:
ZOR filters are STATIC filters with false positives.
-Almost memory optimal: <1% overhead over the theoretical lower bound (!!!)
-Fast queries: ~100 ns
-Construction cannot fail
A thread:
globalprograms.hms.harvard.edu/kalaniyot-hm...
globalprograms.hms.harvard.edu/kalaniyot-hm...
github.com/DerrickWood/... 1/6
github.com/DerrickWood/... 1/6
We introduce new ideas to revisit the notion of sampling with window guarantees, also known as minimizers.
A thread:
We introduce new ideas to revisit the notion of sampling with window guarantees, also known as minimizers.
A thread:
www.biorxiv.org/content/10.1...
This work tackles the issue of Mash which ignores repeats in the genome, providing better distance estimation #GI2025


www.biorxiv.org/content/10.1...
This work tackles the issue of Mash which ignores repeats in the genome, providing better distance estimation #GI2025

If you're a PI working in algorithmic genomics (& you can recommend my lab to your top graduating students ;P), please let them know!
If you're a PI working in algorithmic genomics (& you can recommend my lab to your top graduating students ;P), please let them know!

👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...

👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!

Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!
Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)

Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
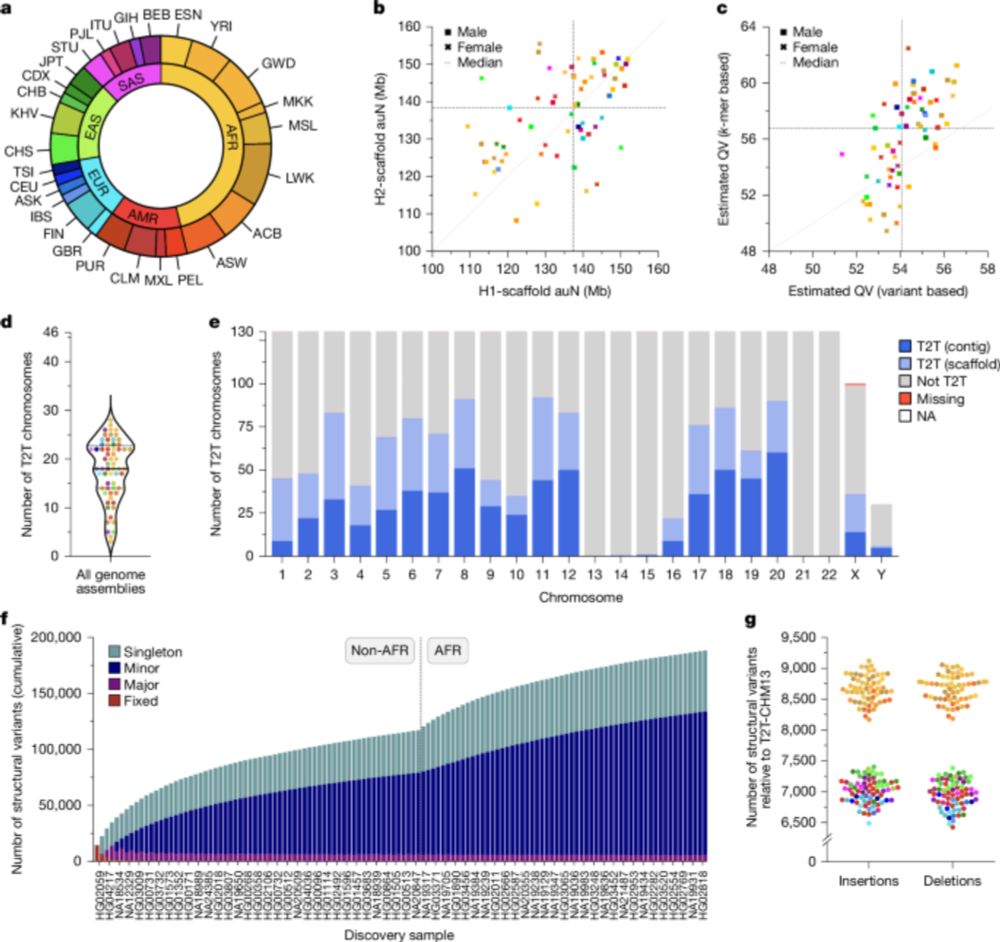
and GH repo: github.com/refresh-bio/...

and GH repo: github.com/refresh-bio/...

