Postdoc in Institut Pasteur in Rayan Chikhi's lab
@martinsteinegger.bsky.social andco
@martinsteinegger.bsky.social andco
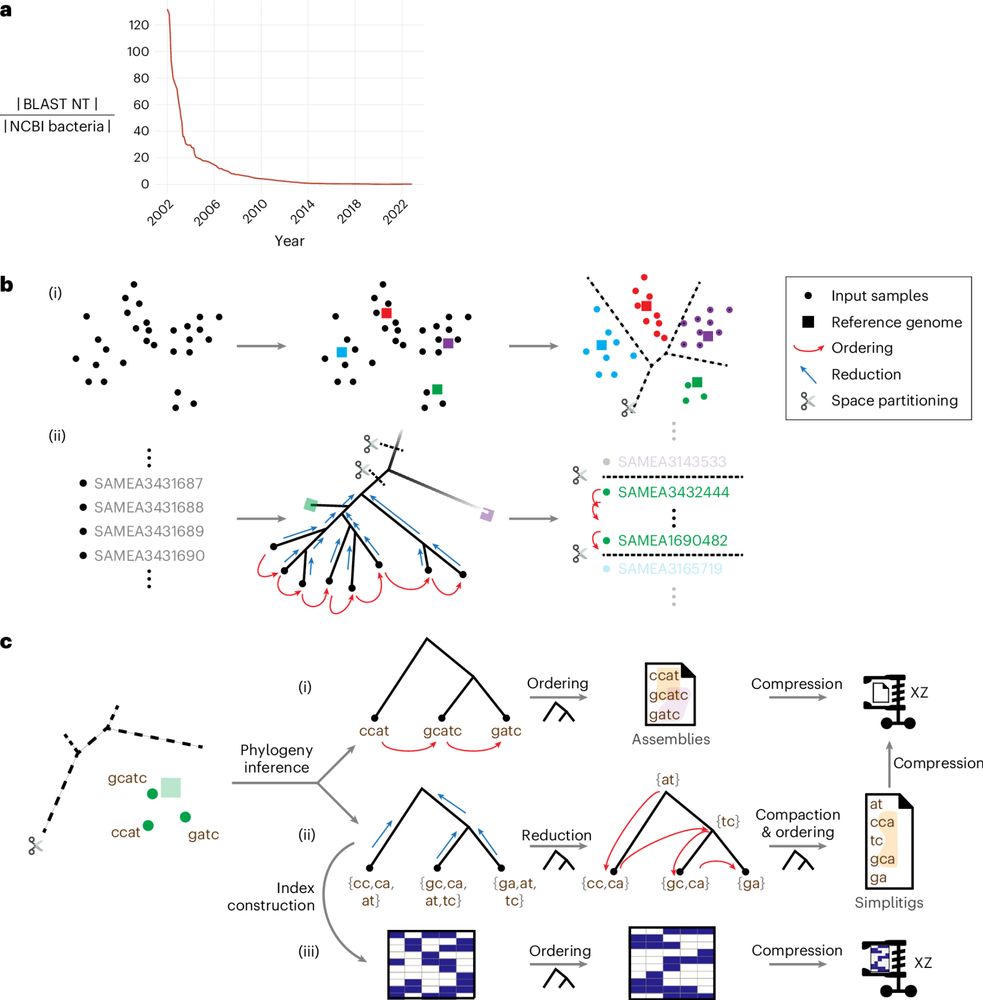
k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu

k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
👉 www.biorxiv.org/content/10.6...

👉 www.biorxiv.org/content/10.6...
We present new strategies to accelerate large-scale document comparison using MinHash-like sketches.
A thread:
We present new strategies to accelerate large-scale document comparison using MinHash-like sketches.
A thread:
👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...

👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
🔗https://www.biorxiv.org/content/10.1101/2025.05.12.653324v1
1/9
🔗https://www.biorxiv.org/content/10.1101/2025.05.12.653324v1
1/9

7/

7/
rdcu.be/eg4OA
1/
If so, you might this little project: github.com/rrwick/conda...
If so, you might this little project: github.com/rrwick/conda...

After discussions with @imartayan.bsky.social, the SIMD minimizer code now also does proper canonical (revcomp) minimizers:
~1ns/bp for fwd minis
+0.4ns/bp with collect and dedup
+0.6ns/bp with canonical hashes.
Super happy how it's only 2x slower in the end!
@yoann.bsky.social
and collaborators.
Reorganize minimizers to allow kmers dichotomic search. That's brilliant.
#bioinformatics 🧬🖥️

@yoann.bsky.social
and collaborators.
Reorganize minimizers to allow kmers dichotomic search. That's brilliant.
#bioinformatics 🧬🖥️
Thanks to my advisors Dominique Lavenier and Jean-François Flot ❤️

Thanks to my advisors Dominique Lavenier and Jean-François Flot ❤️

