Hobby runner 5000m 18:48 | 10k 37:40 | HM 1:27:43 | M 3:39:06

Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
@rayan.chiki.bsky.social
#Bioinformatics


@rayan.chiki.bsky.social
#Bioinformatics


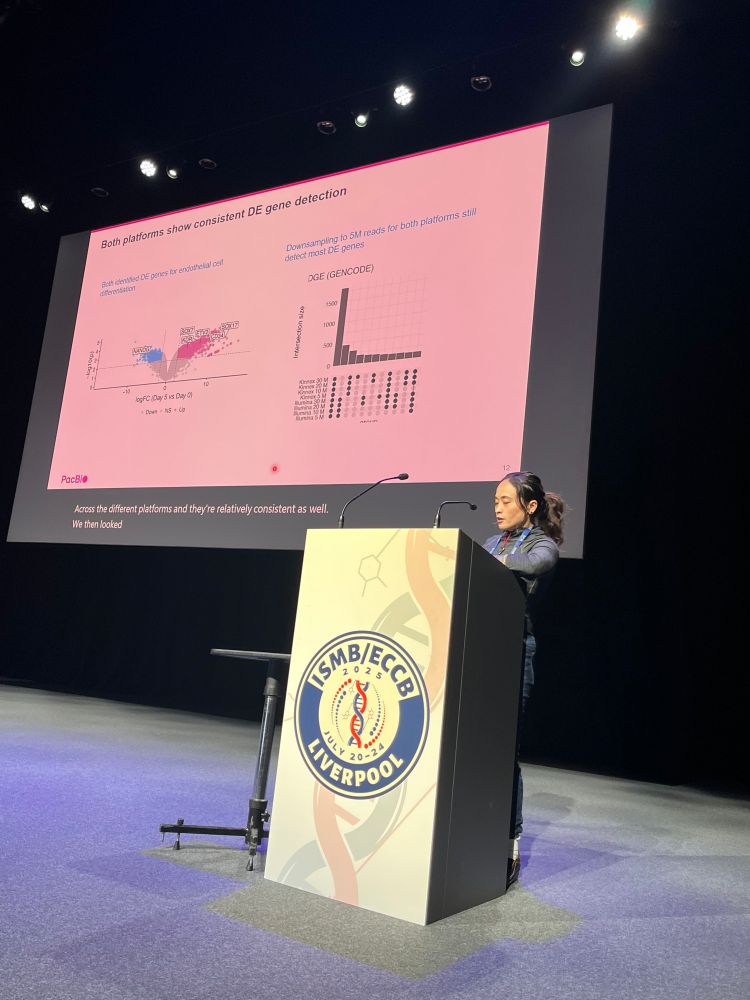
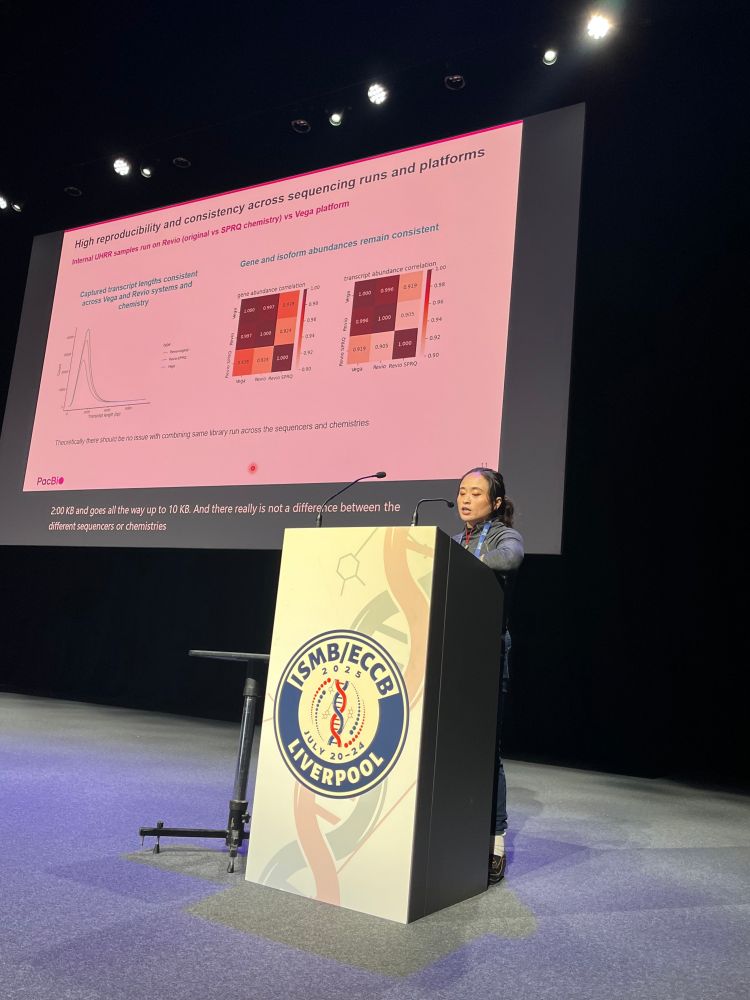
@anaconesa.bsky.social

@anaconesa.bsky.social


Sami is a Doctoral Candidate at @stockholm-uni.bsky.social , working under the supervision of @ksahlin.bsky.social .In this video, Sami shares his research and his role in the broader LongTREC collaboration across Europe.
#AlgorithmDevelopment
Sami is a Doctoral Candidate at @stockholm-uni.bsky.social , working under the supervision of @ksahlin.bsky.social .In this video, Sami shares his research and his role in the broader LongTREC collaboration across Europe.
#AlgorithmDevelopment
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
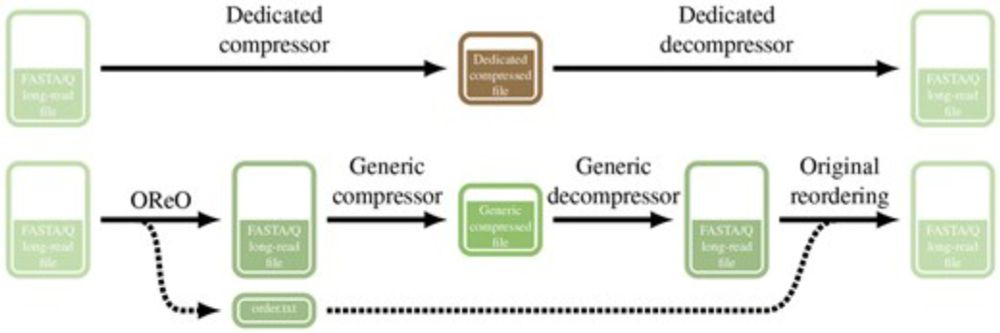
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!

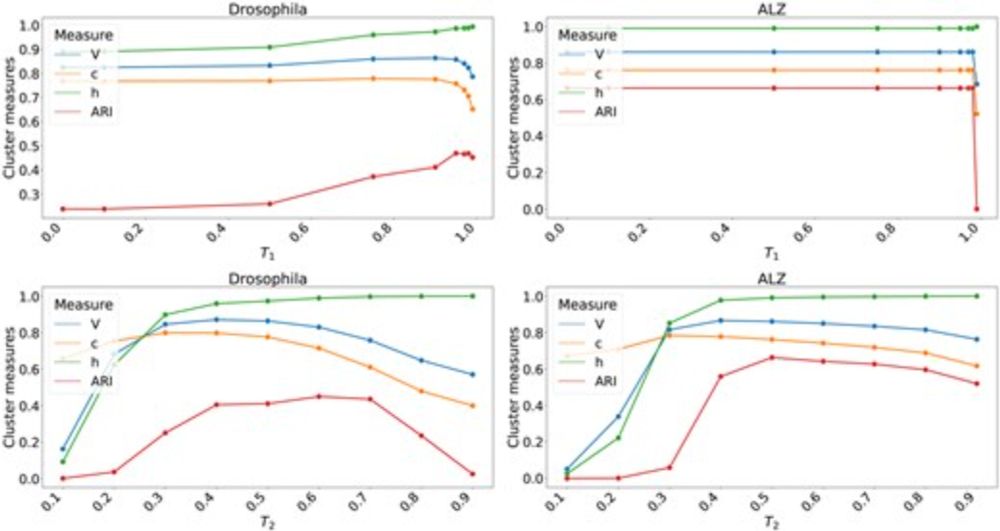
Estimating mutation rates using k-mers is fast—but what happens when repeats dominate the genome?
In a new preprint, Haonan Wu, Antonio Blanca, and myself propose a *repeat-aware* estimator that's accurate even in centromeres.
Estimating mutation rates using k-mers is fast—but what happens when repeats dominate the genome?
In a new preprint, Haonan Wu, Antonio Blanca, and myself propose a *repeat-aware* estimator that's accurate even in centromeres.
O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻

O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻


The last day to submit abstracts for the HitSeq Special Track is April 17th! 🧬
📅 HitSeq is part of ISMB 2025, July 20–24, Liverppol, UK 🇬🇧
📢 Don’t miss your chance to present your work on high-throughput sequencing!
Submit now 👉 www.iscb.org/ismbeccb2025...
#HitSeq #ISMB2025

The last day to submit abstracts for the HitSeq Special Track is April 17th! 🧬
📅 HitSeq is part of ISMB 2025, July 20–24, Liverppol, UK 🇬🇧
📢 Don’t miss your chance to present your work on high-throughput sequencing!
Submit now 👉 www.iscb.org/ismbeccb2025...
#HitSeq #ISMB2025


su.varbi.com/en/what:job/...
Application deadline: April 22. (1/3)

su.varbi.com/en/what:job/...
Application deadline: April 22. (1/3)
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
Fully funded studentships available to work on a range of topics, from small proteins to developing computational tools to study the global microbiome
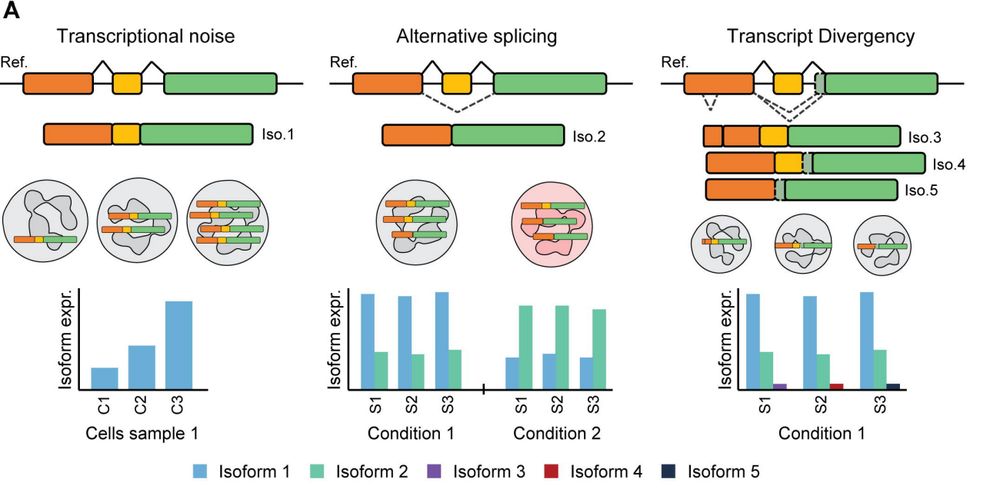
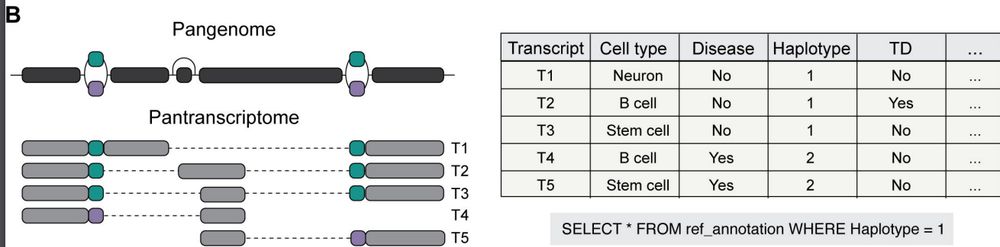
Are you passionate about:
🧬 Graph algorithms for real-world genome sequencing?
💻 Writing efficient, reusable code & libraries?
🌲 Exploring stunning Nordic nature?
This PhD position is for YOU! 🎓✨
📅 Apply by March 2
#PhD #ComputerScience #Bioinformatics #GraphAlgorithms
Are you passionate about:
🧬 Graph algorithms for real-world genome sequencing?
💻 Writing efficient, reusable code & libraries?
🌲 Exploring stunning Nordic nature?
This PhD position is for YOU! 🎓✨
📅 Apply by March 2
#PhD #ComputerScience #Bioinformatics #GraphAlgorithms
Low mem construction. Spaced seeds. Optional crazy-low-mem search. More info on the horizon.
crates.io/crates/sufr

Low mem construction. Spaced seeds. Optional crazy-low-mem search. More info on the horizon.


