Silesian University of Technology
bioinformatician, computer scientist

and GH repo: github.com/refresh-bio/...

and GH repo: github.com/refresh-bio/...
@uni-jena.de @microverse.bsky.social
www.nature.com/articles/s41...

@uni-jena.de @microverse.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Great collaboration with A.Zielezinki, UAM guys and @bedutilh.bsky.social

www.nature.com/articles/s41...
Great collaboration with A.Zielezinki, UAM guys and @bedutilh.bsky.social
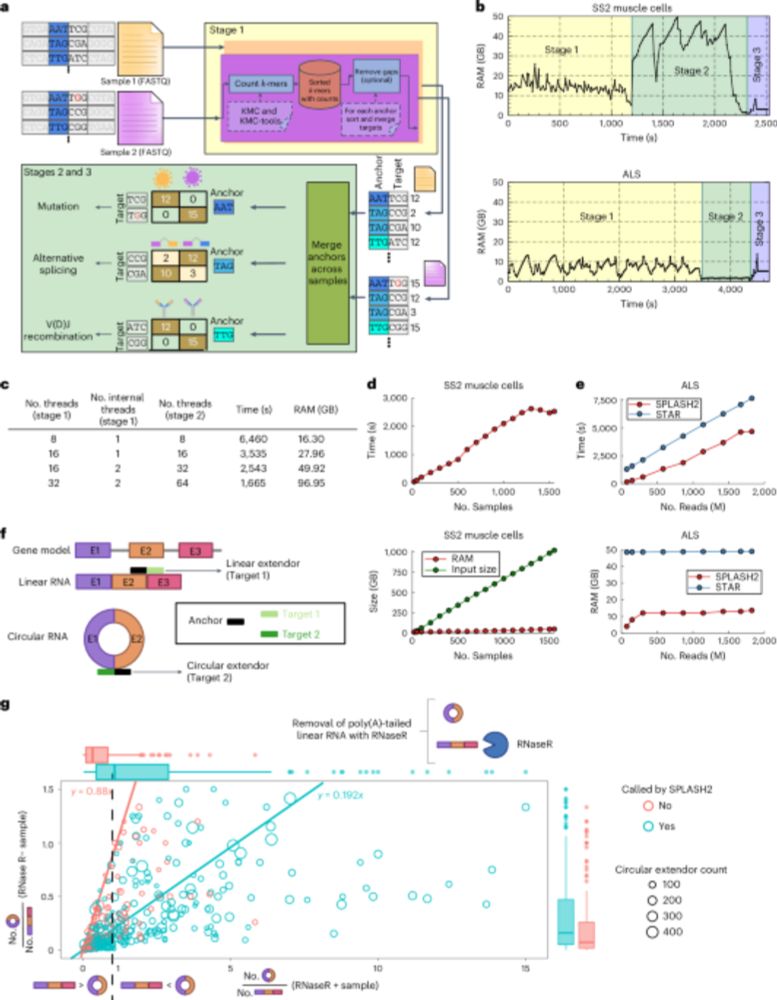
github.com/refresh-bio/...
github.com/refresh-bio/...

