We discovered a new NAD⁺-depleting bacterial immune system aRES and phage enzymes that overcome it.
Our preprint is out: www.biorxiv.org/content/10.6...
Avs5 detects an early jumbo‑phage activator and halts infection by rapidly hydrolyzing NAD+.
www.sciencedirect.com/science/arti... #phage #bacteriophage #MicroSky

Avs5 detects an early jumbo‑phage activator and halts infection by rapidly hydrolyzing NAD+.
www.sciencedirect.com/science/arti... #phage #bacteriophage #MicroSky
www.nature.com/articles/s41...

www.nature.com/articles/s41...
A pore-forming antiphage defence is activated by oligomeric phage proteins
-from Maxwell & Norris
www.nature.com/articles/s41...
Bacterial immune activation via supramolecular assembly with phage triggers
-from Laub & Ghanbarpour
www.nature.com/articles/s41...
A pore-forming antiphage defence is activated by oligomeric phage proteins
-from Maxwell & Norris
www.nature.com/articles/s41...
Bacterial immune activation via supramolecular assembly with phage triggers
-from Laub & Ghanbarpour
www.nature.com/articles/s41...
Small molecules inhibit type II Thoeris anti-phage systems from diverse bacteria. One compound, IP6C, improves phage-therapy against P. aeruginosa & is effective against Thoeris in polymicrobial communities
www.cell.com/cell-host-mi...

Small molecules inhibit type II Thoeris anti-phage systems from diverse bacteria. One compound, IP6C, improves phage-therapy against P. aeruginosa & is effective against Thoeris in polymicrobial communities
www.cell.com/cell-host-mi...
📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...

📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...
We discovered a new NAD⁺-depleting bacterial immune system aRES and phage enzymes that overcome it.
Our preprint is out: www.biorxiv.org/content/10.6...
We discovered a new NAD⁺-depleting bacterial immune system aRES and phage enzymes that overcome it.
Our preprint is out: www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
Asgard archaea and the origin of eukaryotes! 🦠
Eukaryotic cellular complexity evolved largely within the Asgard lineage before mitochondrial endosymbiosis and later bacterial gene acquisitions
#MicroSky
www.nature.com/articles/s41...

Asgard archaea and the origin of eukaryotes! 🦠
Eukaryotic cellular complexity evolved largely within the Asgard lineage before mitochondrial endosymbiosis and later bacterial gene acquisitions
#MicroSky
www.nature.com/articles/s41...

A new study from the Alex Gao lab. The scope of this work is incredible!!!
www.biorxiv.org/content/10.6...

A new study from the Alex Gao lab. The scope of this work is incredible!!!
www.biorxiv.org/content/10.6...
journals.asm.org/doi/10.1128/...

journals.asm.org/doi/10.1128/...

www.nature.com/articles/s41...
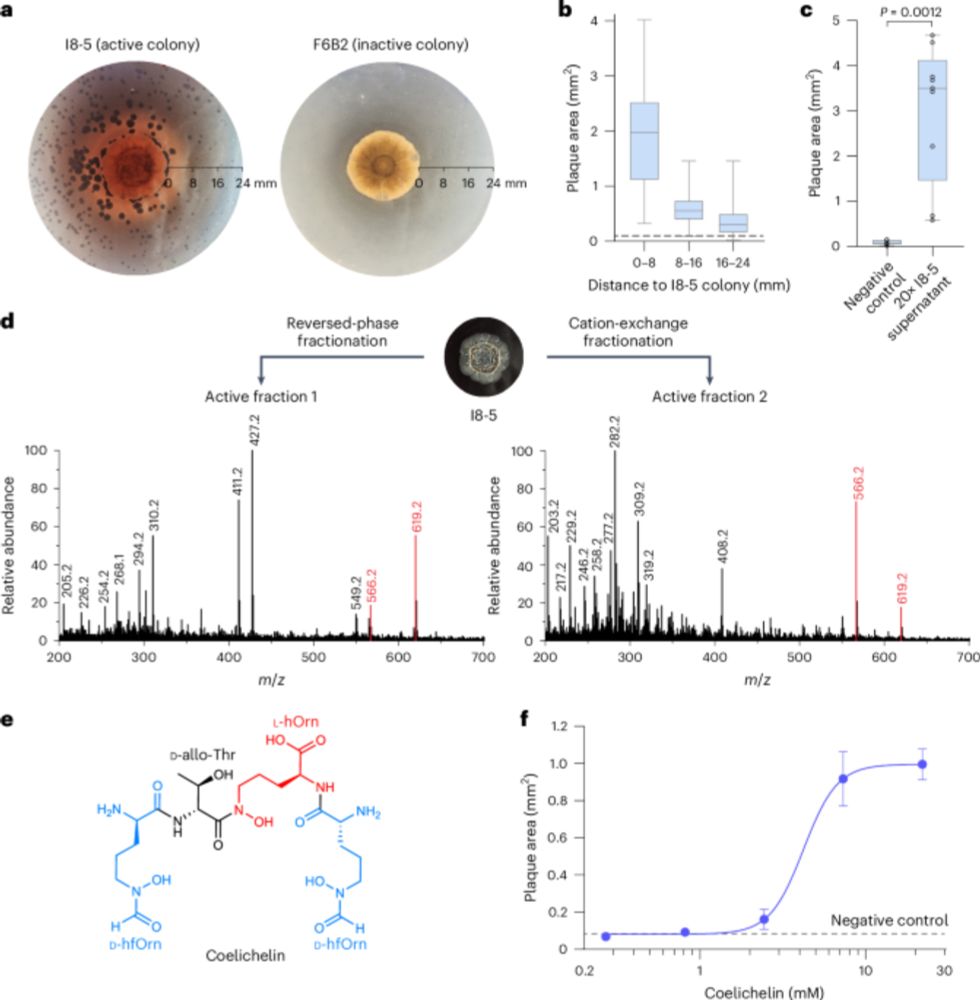
www.nature.com/articles/s41...
➡️ We co-evolved two bacterial strains in conditions in which the costs and benefits of prophage carriage varied
Here is what we found. www.biorxiv.org/content/10.6...
#MicroSky #PhageSky
🧵

➡️ We co-evolved two bacterial strains in conditions in which the costs and benefits of prophage carriage varied
Here is what we found. www.biorxiv.org/content/10.6...
#MicroSky #PhageSky
🧵
Phage proteins inhibit bacterial transcription. Novel inhibitors binding RNA polymerase discovered via structural predictions.




Phage proteins inhibit bacterial transcription. Novel inhibitors binding RNA polymerase discovered via structural predictions.

New preprint from the lab !!! 🥳
In this work, we explore the 3D genome architecture of the virulent phage PAK_P3 during its infection cycle in P. Aeruginosa. We unveil a highly dynamic structuration as well as specific interactions patterns.
www.biorxiv.org/content/bior...
New preprint from the lab !!! 🥳
In this work, we explore the 3D genome architecture of the virulent phage PAK_P3 during its infection cycle in P. Aeruginosa. We unveil a highly dynamic structuration as well as specific interactions patterns.
www.biorxiv.org/content/bior...
In this preprint, Lucas Paoli et al, ask what shapes antiphage systems expression in native contexts.
www.biorxiv.org/content/10.6...

In this preprint, Lucas Paoli et al, ask what shapes antiphage systems expression in native contexts.
www.biorxiv.org/content/10.6...
Igor Fesenko, Svetlana A Shabalina, Gisela Storz, Eugene V Koonin.
Nucleic Acids Research, Volume 53, Issue 22, 11 December 2025
doi.org/10.1093/nar/...
Igor Fesenko, Svetlana A Shabalina, Gisela Storz, Eugene V Koonin.
Nucleic Acids Research, Volume 53, Issue 22, 11 December 2025
doi.org/10.1093/nar/...


