Join us, collaborate or learn about what we do:
mancinilab.github.io/index.html
And check out our most recent output:
www.biorxiv.org/content/10.1...
Join us, collaborate or learn about what we do:
mancinilab.github.io/index.html
And check out our most recent output:
www.biorxiv.org/content/10.1...
Big thanks to James for presenting his work on advanced CRISPR tools for epitranscriptomic research, and to @eu-life.bsky.social for this opportunity!🧬✈️
#EULIFE #CRG

Big thanks to James for presenting his work on advanced CRISPR tools for epitranscriptomic research, and to @eu-life.bsky.social for this opportunity!🧬✈️
#EULIFE #CRG
Big thanks to the organisers for such an inspiring and fun event 👏
Congrats to @miemonti.bsky.social for being part of the organising team and to @oguzhanbegik.bsky.social for giving a fantastic talk! 🎤✨
#GenomeBiology #CRG #NovoaLab




Big thanks to the organisers for such an inspiring and fun event 👏
Congrats to @miemonti.bsky.social for being part of the organising team and to @oguzhanbegik.bsky.social for giving a fantastic talk! 🎤✨
#GenomeBiology #CRG #NovoaLab




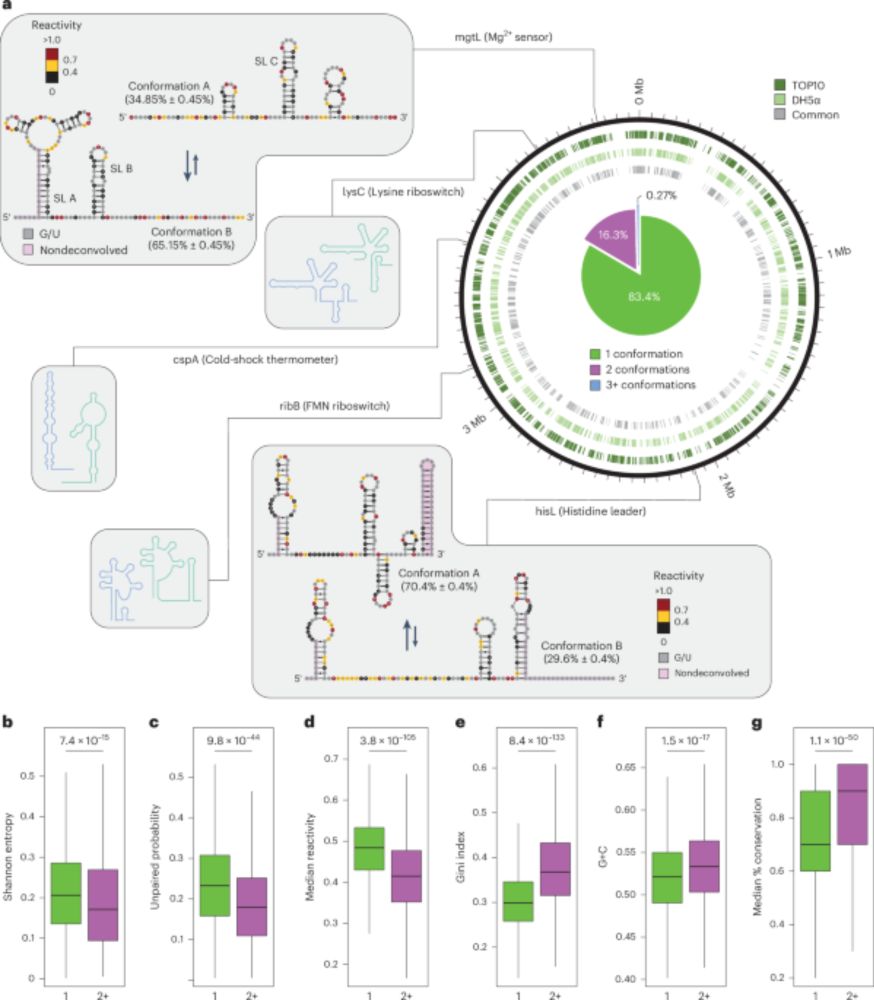
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)

We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)
Mechanobiology meets Nuclear Metabolism
www.nature.com/articles/s41...
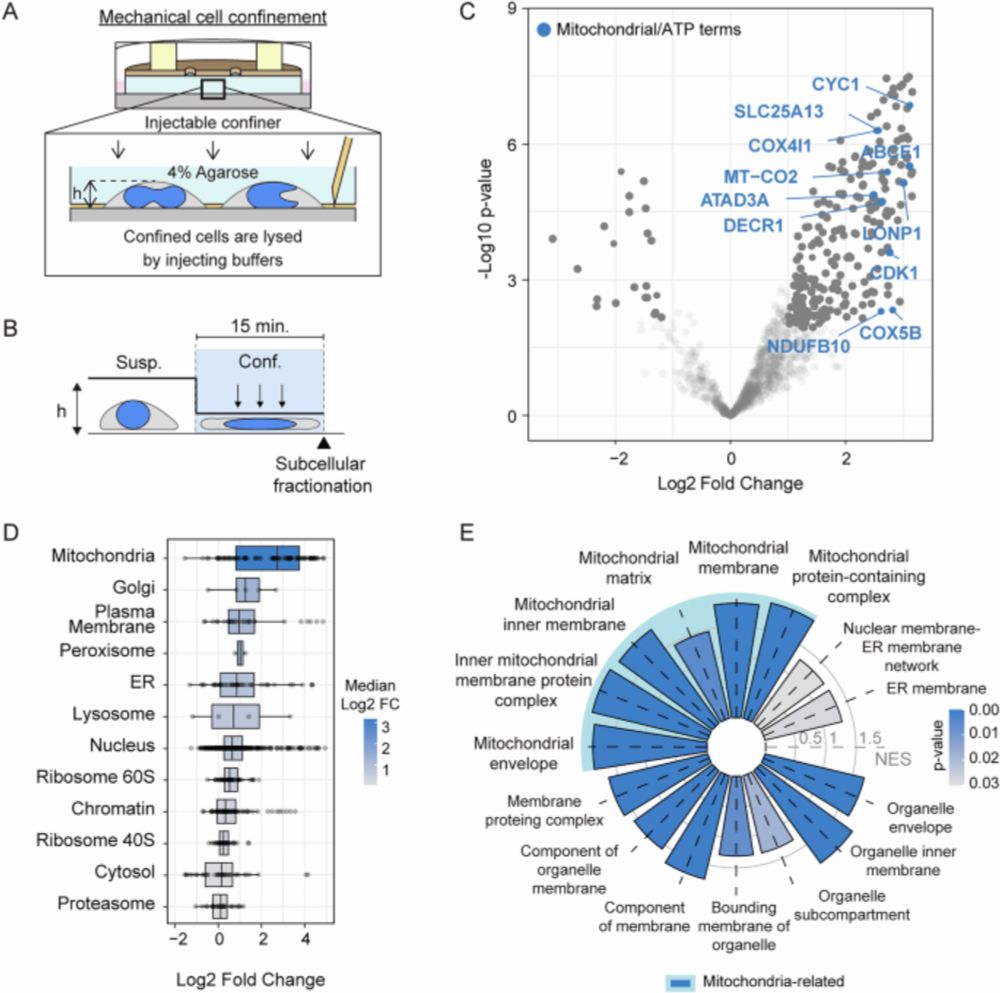
Mechanobiology meets Nuclear Metabolism
www.nature.com/articles/s41...
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

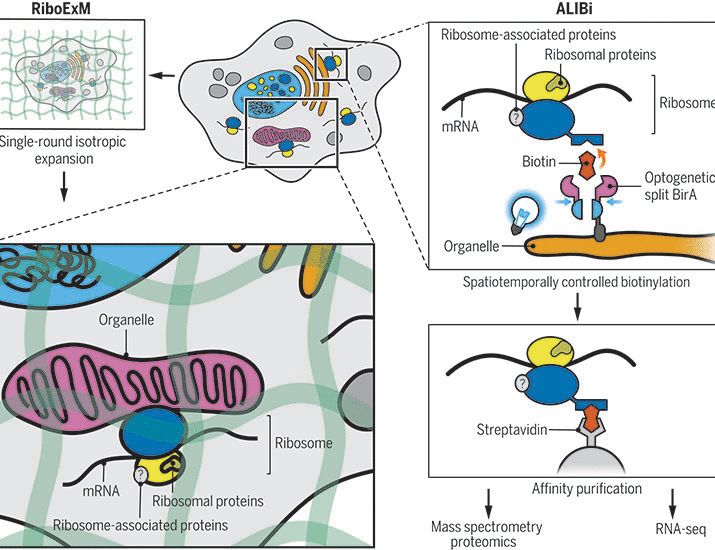
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)
Huge congratulations again, Eva - an incredible and well-deserved recognition! 🧬🏆



Huge congratulations again, Eva - an incredible and well-deserved recognition! 🧬🏆
A big thank you to everyone who made it such a success — from the science-packed discussions and hands-on workshops to the brainstorming, games, sunny pool sessions, and delicious BBQs. And special thanks to the organizers!⚘️
@evamarianovoa.bsky.social




www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...

"While the minimum basis for a thriving European Research & Innovation is a stand-alone, well budgeted #FP10, we urge the European Commission to further follow 5 key actions"
lnkd.in/d2mzN_zu
"While the minimum basis for a thriving European Research & Innovation is a stand-alone, well budgeted #FP10, we urge the European Commission to further follow 5 key actions"
lnkd.in/d2mzN_zu
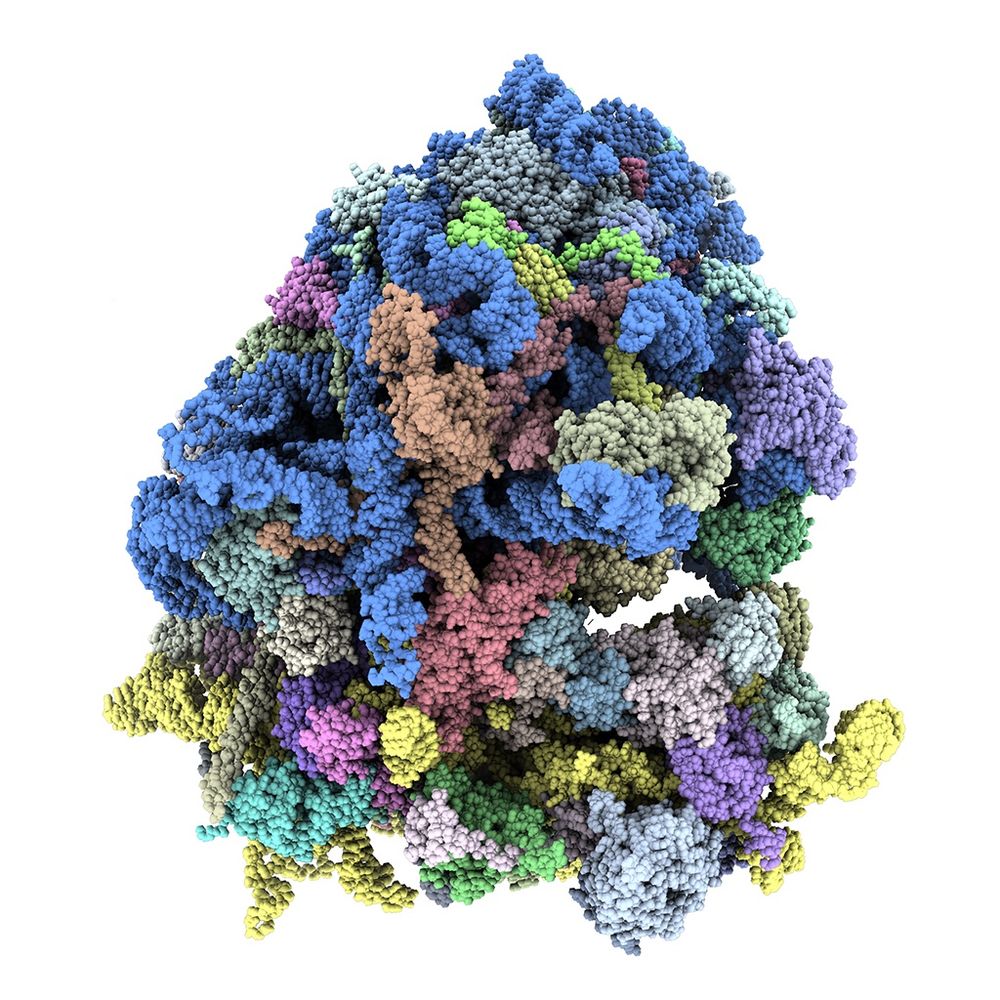
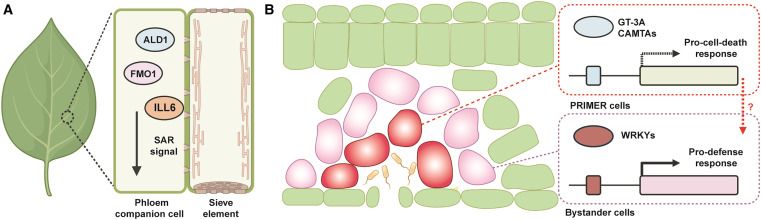

A 🧵 below 👇🏼