github.com/mfansler
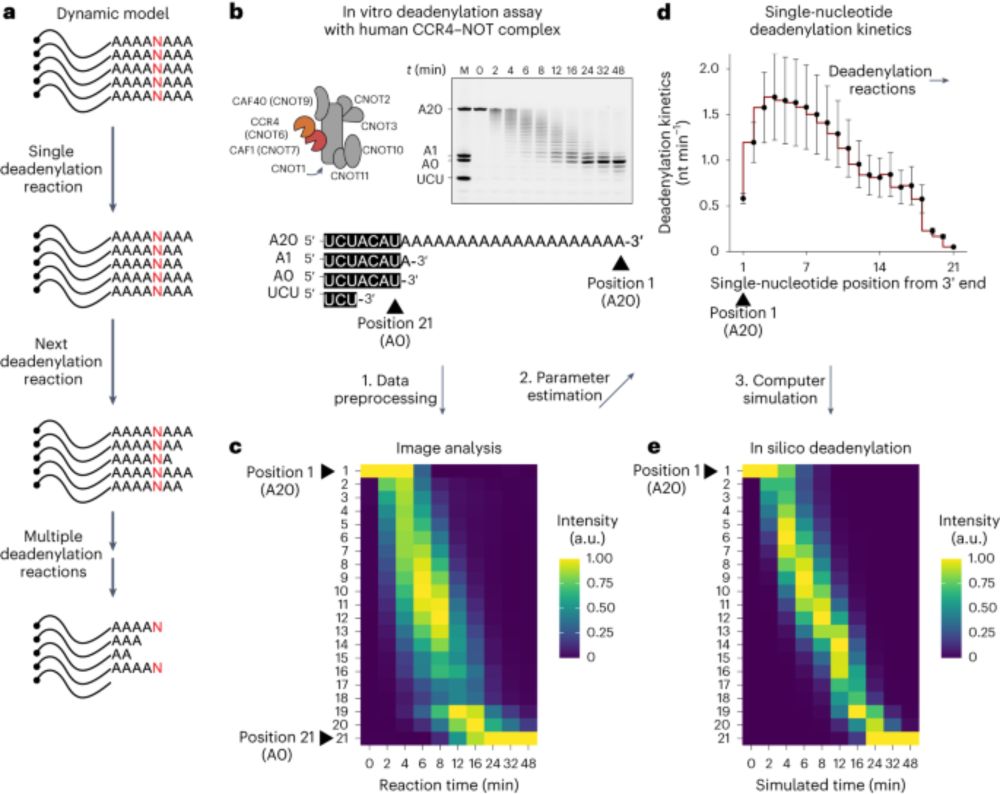
Delighted to share the story of two germline RBPs - one with little (DND1) and one with no (NANOS3) intrinsic sequence-specificity - that together build a continuous RNA binding surface recognizing a 7-mer (AUGAAUU) in target mRNA 3’UTRs, leading to deadenylation.

Delighted to share the story of two germline RBPs - one with little (DND1) and one with no (NANOS3) intrinsic sequence-specificity - that together build a continuous RNA binding surface recognizing a 7-mer (AUGAAUU) in target mRNA 3’UTRs, leading to deadenylation.
One can say that much of the progress in computer programming is aligned with ever increasing levels abstraction.
LLM-based coding tools are "just" a next level.
The choice of objective, and the measure of success, still remains something external to the tool.
One can say that much of the progress in computer programming is aligned with ever increasing levels abstraction.
LLM-based coding tools are "just" a next level.
The choice of objective, and the measure of success, still remains something external to the tool.
Come to EMBL! Scope includes physics-based models, emergence, statistics, ML/AI
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
Come to EMBL! Scope includes physics-based models, emergence, statistics, ML/AI
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
Also see linking LAPACK, when only BLAS needed, but a bit more understandable.
Also see linking LAPACK, when only BLAS needed, but a bit more understandable.
some think this field, despite recent Nobel to Ambros and Ruvkun, doesn't have much surprises left. but, do we EVEN KNOW WHAT a miRNA precursor LOOKS LIKE? we found some conserved miRNA hairpins up to 10x longer than suspected. this is mir-12, just bonkers! 1/n

some think this field, despite recent Nobel to Ambros and Ruvkun, doesn't have much surprises left. but, do we EVEN KNOW WHAT a miRNA precursor LOOKS LIKE? we found some conserved miRNA hairpins up to 10x longer than suspected. this is mir-12, just bonkers! 1/n
It's not every day we have a new major #ggplot2 release but it is a fitting 18 year birthday present for the package.
Get an overview of the release in this blog post and be on the lookout for more in-depth posts #rstats

github.com/NPSDC/mehenDi
github.com/NPSDC/mehenDi
"Rethinking RNA-binding proteins: Riboregulation challenges prevailing views" www.cell.com/cell/fulltex...

"Rethinking RNA-binding proteins: Riboregulation challenges prevailing views" www.cell.com/cell/fulltex...

Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
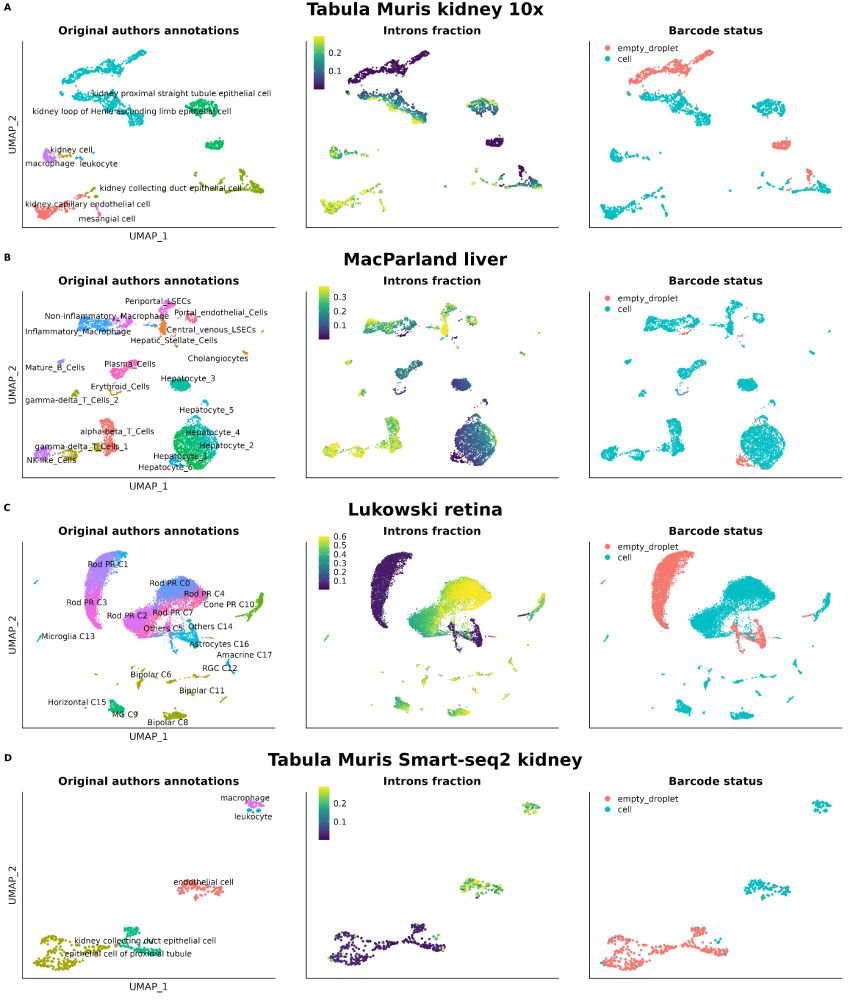
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
www.nature.com/articles/s41...
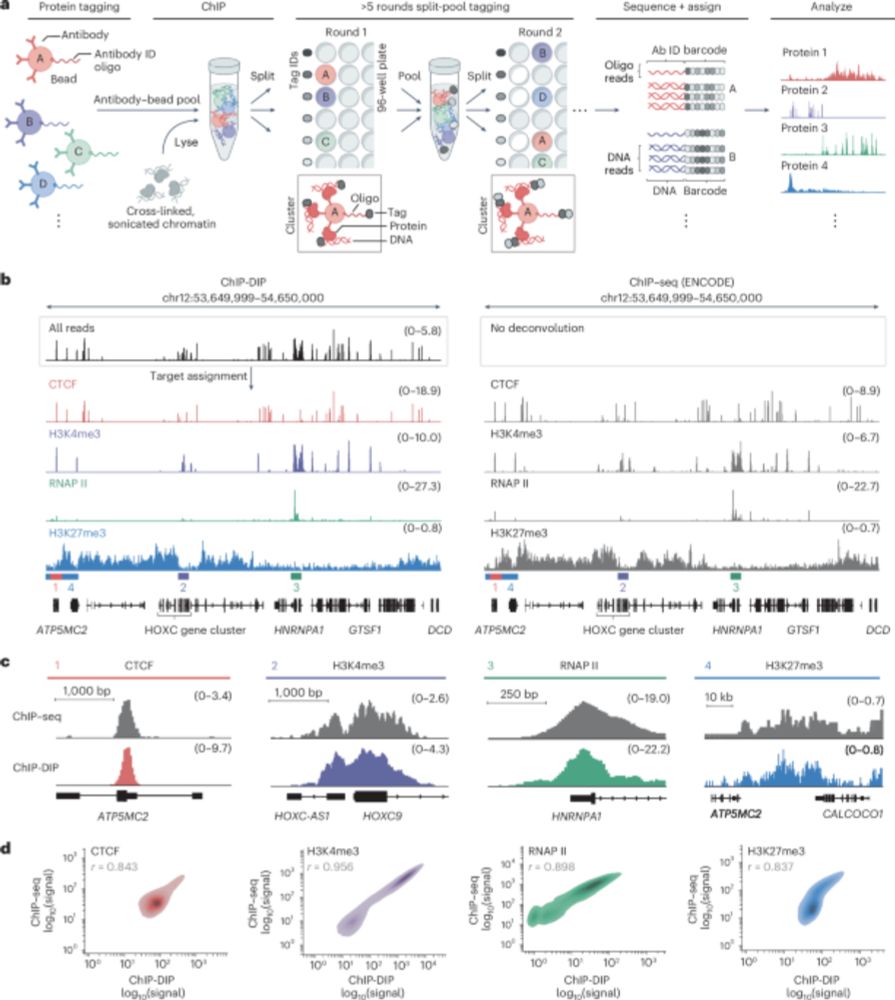
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
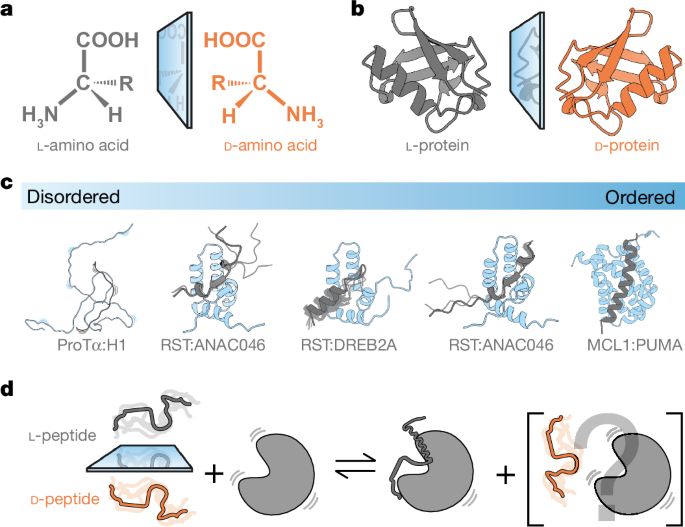
www.nature.com/articles/s41...

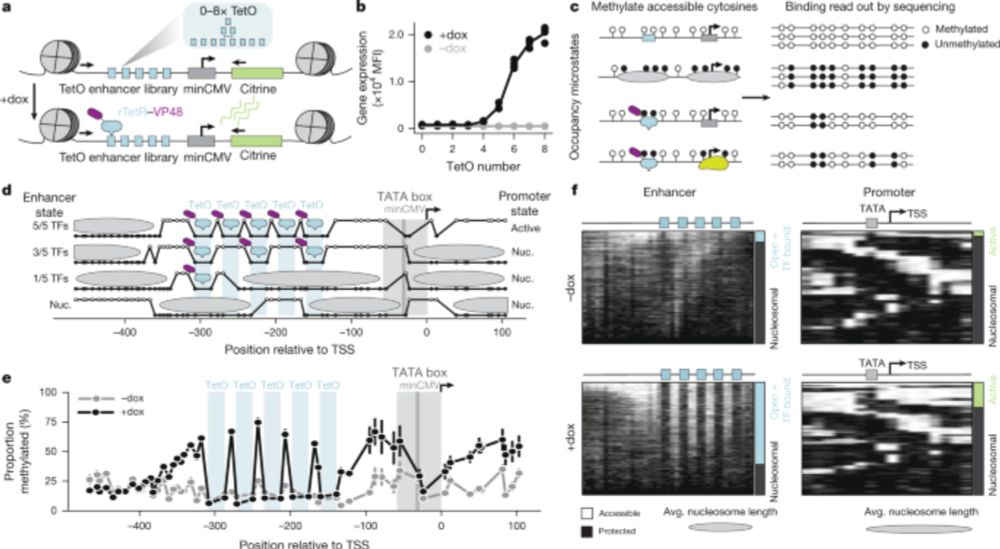
Develops SCimilarity which they use to query 23.4M cell atlas to find similar cell profiles.
#Genomics #SingleCell
www.nature.com/articles/s41...

Develops SCimilarity which they use to query 23.4M cell atlas to find similar cell profiles.
#Genomics #SingleCell
www.nature.com/articles/s41...

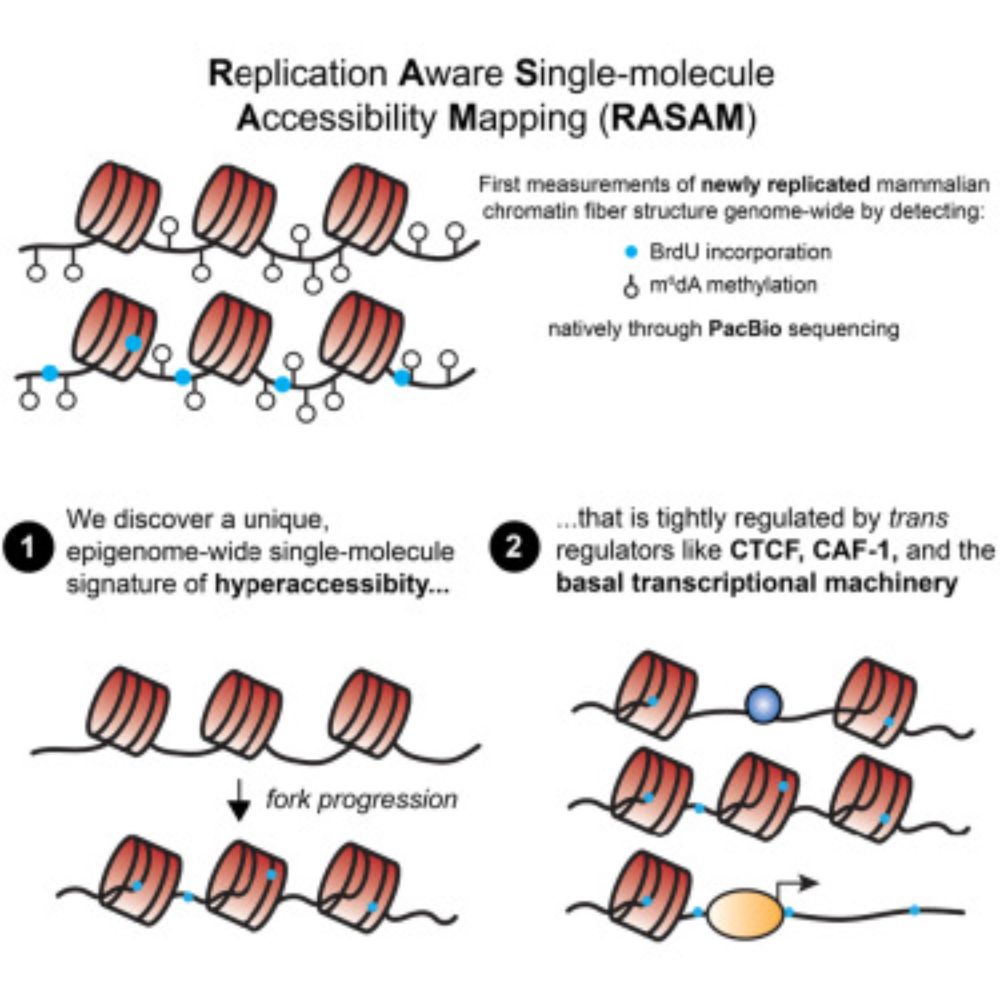

github.com/mfansler

github.com/mfansler