

DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
DeGrado, Arikin, Picotti (Keynotes). @lmkdassama.bsky.social @brianliau.bsky.social @rhodamine110.bsky.social @benlehner.bsky.social @alitavassoli.bsky.social
www.embl.org/about/info/c...
https://blog.stephenturner.us/p/posit-conf-2025-youtube-playlist
#rstats #datascience

https://blog.stephenturner.us/p/posit-conf-2025-youtube-playlist
#rstats #datascience
⬇️
academic.oup.com/nar/article/...

⬇️
academic.oup.com/nar/article/...
Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
🧬💻 #MedSky
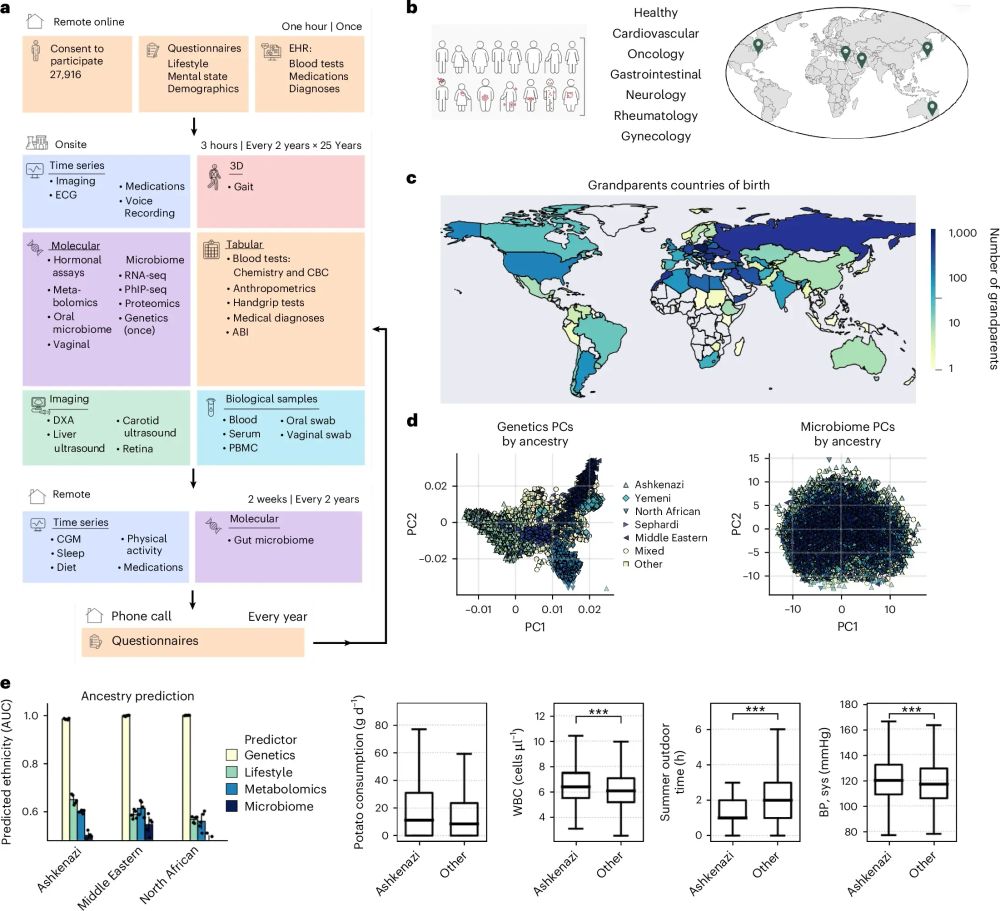
🧬💻 #MedSky
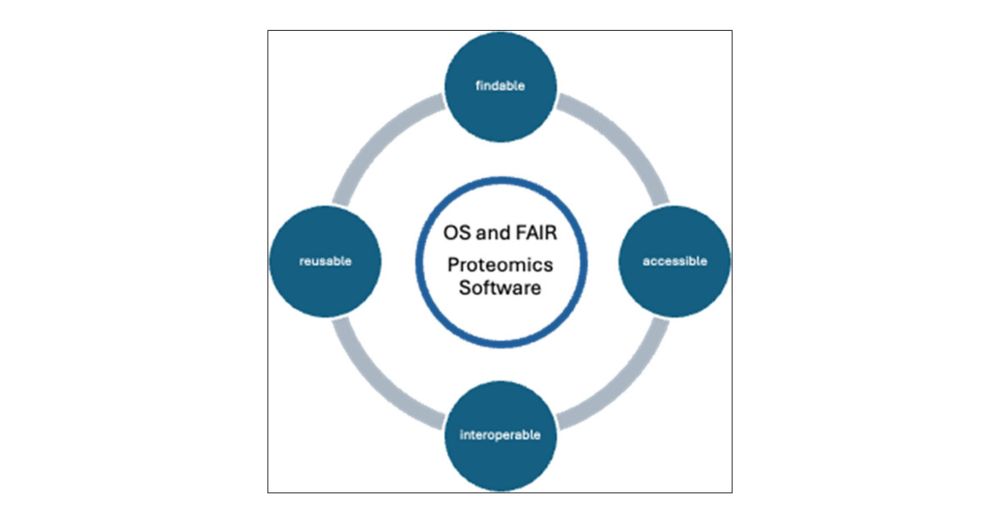
Meet the Team and Labs behind the project
👉 quantms.org/about
#OpenSource #Proteomics #ASMS2025
Meet the Team and Labs behind the project
👉 quantms.org/about
#OpenSource #Proteomics #ASMS2025
marimo-team.github.io/quarto-marimo/
What's marimo? Reactive #jupyter -like notebooks that automatically run cells when code changes. marimo islands is making this tech portable everywhere 🏝️
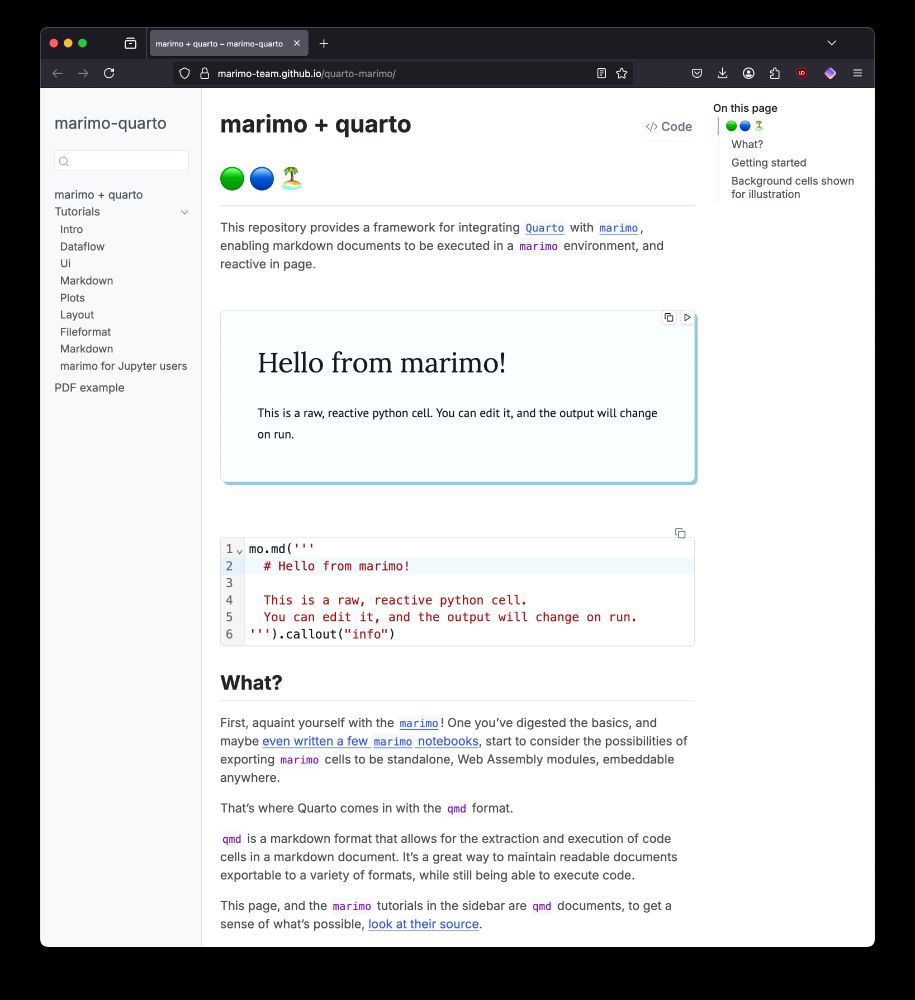
marimo-team.github.io/quarto-marimo/
What's marimo? Reactive #jupyter -like notebooks that automatically run cells when code changes. marimo islands is making this tech portable everywhere 🏝️
doi.org/10.1039/D5SC...
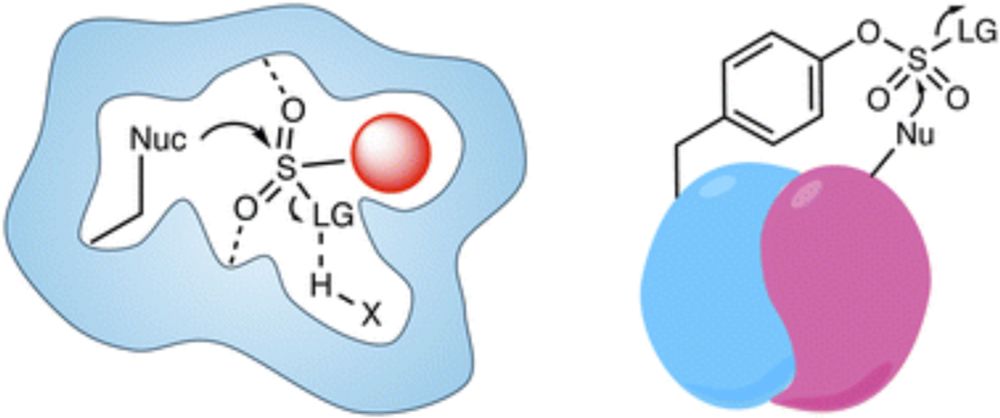
doi.org/10.1039/D5SC...
---
#proteomics #prot-paper
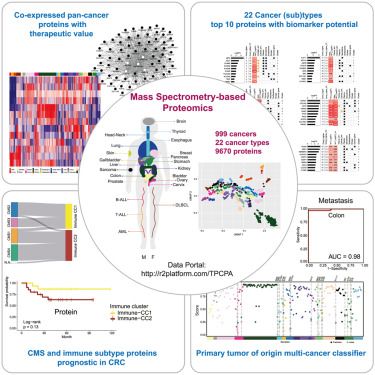
---
#proteomics #prot-paper
www.embopress.org/doi/full/10....

www.embopress.org/doi/full/10....
pubs.acs.org/doi/10.1021/...
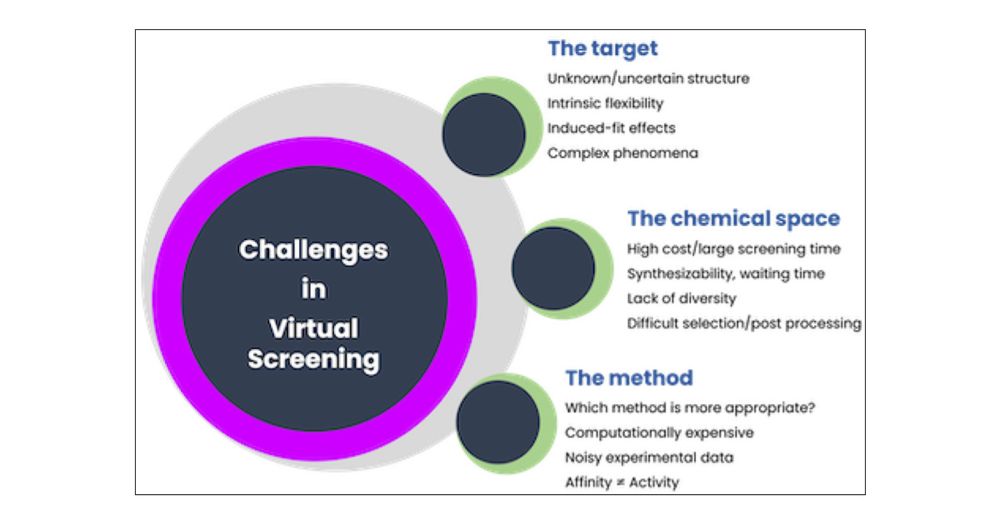
pubs.acs.org/doi/10.1021/...

#CompChem #ML 🧪 ⚗️

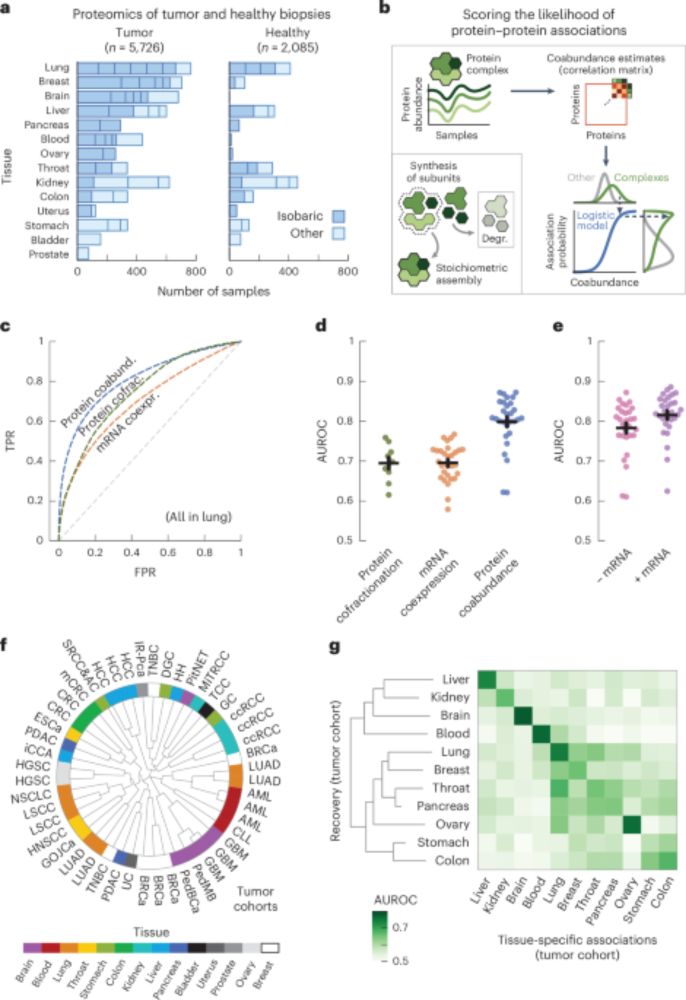
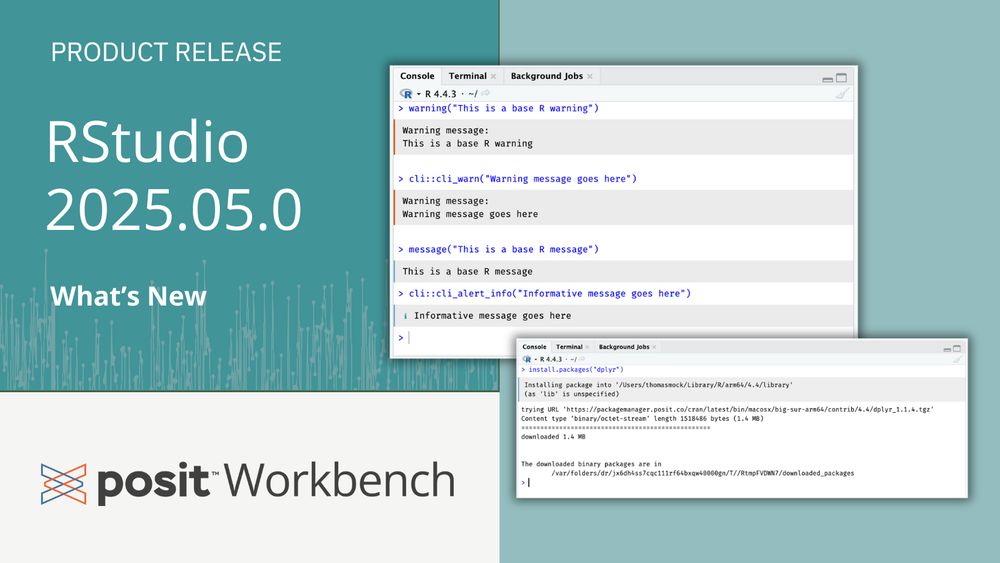
The latest release includes Quarto 1.6 support, genAI integrations, better errors/warnings in the console, and other improvements.
Read more in the blog post: posit.co/blog/rstudio...
#RStats #Python
The latest release includes Quarto 1.6 support, genAI integrations, better errors/warnings in the console, and other improvements.
Read more in the blog post: posit.co/blog/rstudio...
#RStats #Python
- Smarter chat responses with new tools
- Improved multi-window support
- Image and Streamable HTTP support for MCP servers
…and so much more. aka.ms/VSCodeRelease
Here are some of the highlights… 🧵
![A thumbnail with a dark background and the VS Code logo that reads “What’s new in Visual Studio Code April Update [1.100] -Performance improvements in chat -Auto-attach instructions to chat -Reuse prompts for common tasks -Smarter chat responses with GitHub, extensions, and notebook tools -Identify staged changes in editor -Improved multi-window support -Quick create for Python Environments -Image and Streamable HTTP support for MCP servers”](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2xibtqlq4oplg5hkbqw56xgu/bafkreih3cfvsxjemy3ni2symoe77ag7tbbarx77wn4e6uzwzae6eztk73y@jpeg)
- Smarter chat responses with new tools
- Improved multi-window support
- Image and Streamable HTTP support for MCP servers
…and so much more. aka.ms/VSCodeRelease
Here are some of the highlights… 🧵
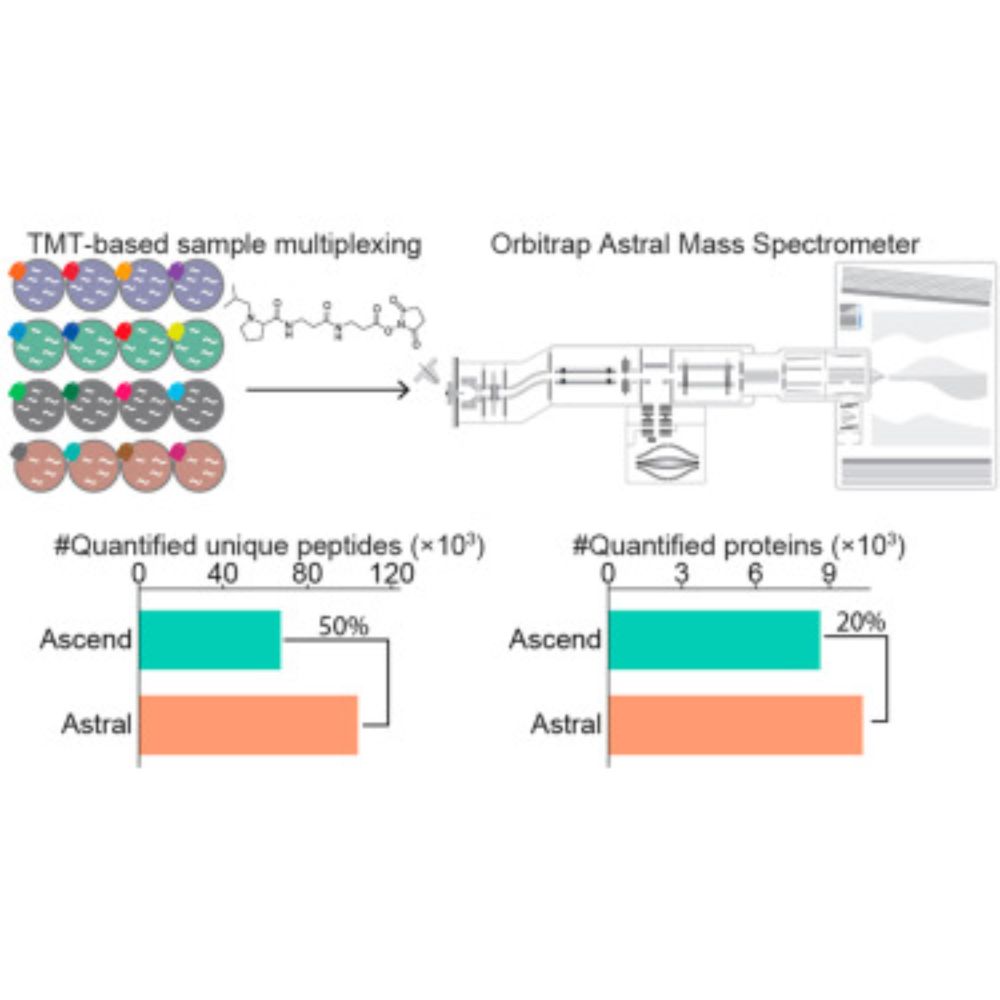
🔗 Supplementary material: ars.els-cdn.com/content/imag...
#ChemicalProbes #DrugDiscovery #ChemicalBiology
🔗 Supplementary material: ars.els-cdn.com/content/imag...
#ChemicalProbes #DrugDiscovery #ChemicalBiology
#proteomics #metabolomics #lipidomics
discord.gg/Sm6gWgpsf4

authors.elsevier.com/c/1koBJL7PXu...

authors.elsevier.com/c/1koBJL7PXu...
#SBDD predictions need reliable benchmarks - diverse targets, high-quality affinity & structural data, and blinded validation. Let’s make it happen!
🔗 Read more: doi.org/10.1021/acs....
#DrugDiscovery #CompChem
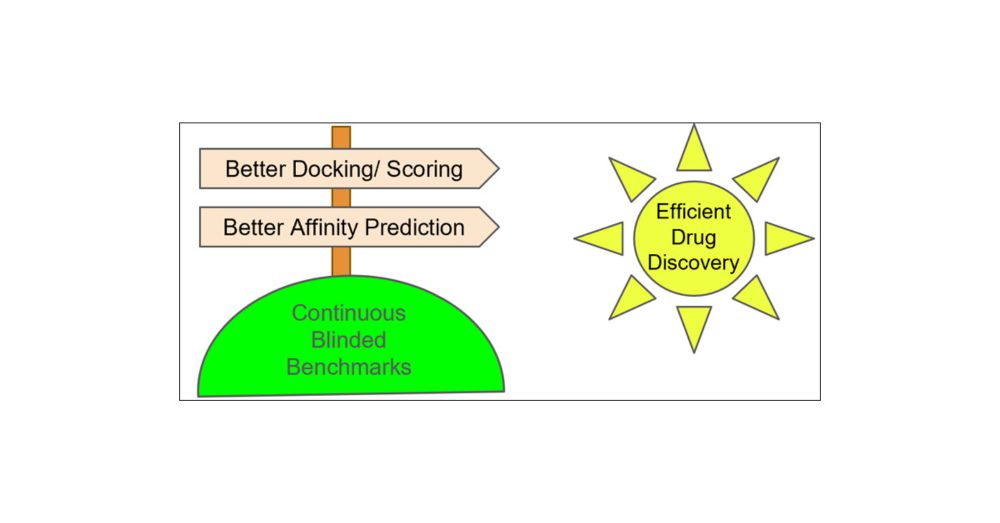
#SBDD predictions need reliable benchmarks - diverse targets, high-quality affinity & structural data, and blinded validation. Let’s make it happen!
🔗 Read more: doi.org/10.1021/acs....
#DrugDiscovery #CompChem

