johahi.github.io
We address this in our preprint “Aberrant splicing prediction during human organ development”: www.biorxiv.org/content/10.1...
We address this in our preprint “Aberrant splicing prediction during human organ development”: www.biorxiv.org/content/10.1...
👉 AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model
📅 Wed Feb 4, 5:30pm CET
🧬 kipoi.org/seminar
🦋 @kipoizoo.bsky.social
👉 AlphaGenome: advancing regulatory variant effect prediction with a unified DNA sequence model
📅 Wed Feb 4, 5:30pm CET
🧬 kipoi.org/seminar
🦋 @kipoizoo.bsky.social
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Poster 5022W, Wed 2:30-4:30.

Poster 5022W, Wed 2:30-4:30.
We enhanced Borzoi with RoPE & FlashAttention for >3x faster training/inference & 2.4x reduction in memory usage.
This brings large-scale genomic analysis and fine-tuning within reach of academic budgets.
📄: doi.org/10.1093/bioi...

We enhanced Borzoi with RoPE & FlashAttention for >3x faster training/inference & 2.4x reduction in memory usage.
This brings large-scale genomic analysis and fine-tuning within reach of academic budgets.
📄: doi.org/10.1093/bioi...
www.nature.com/articles/s41...
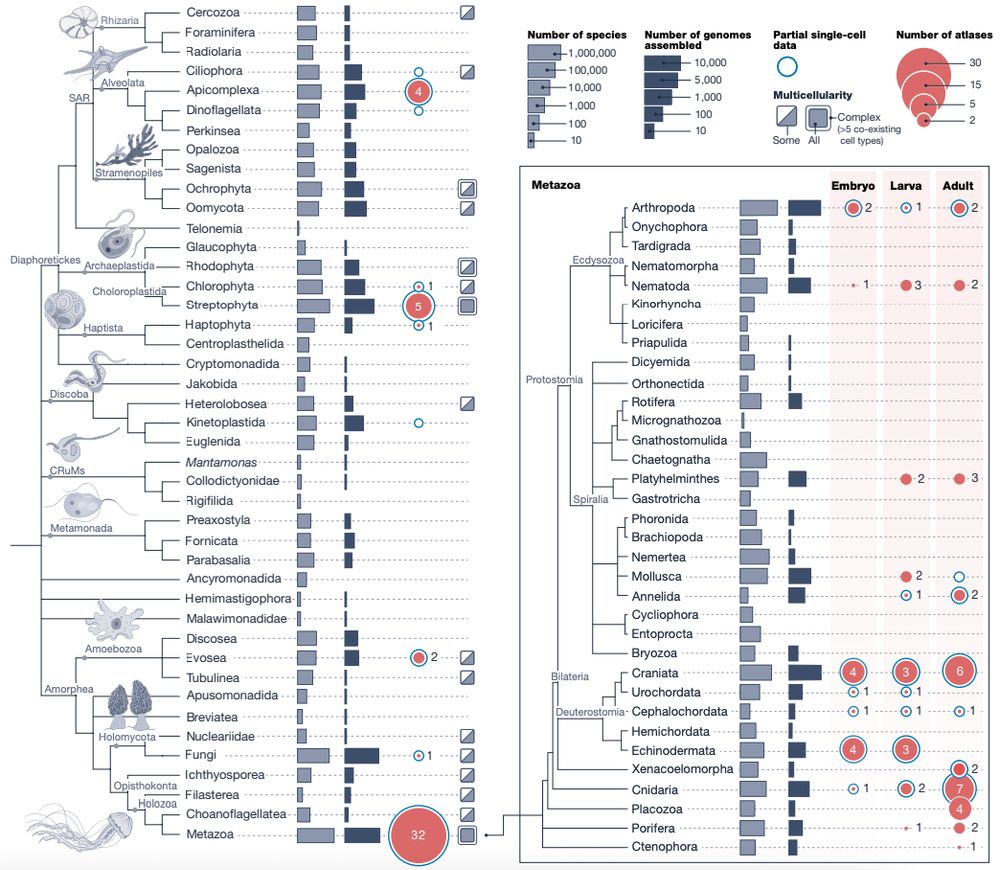
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
doi.org/10.1101/2025...

doi.org/10.1101/2025...
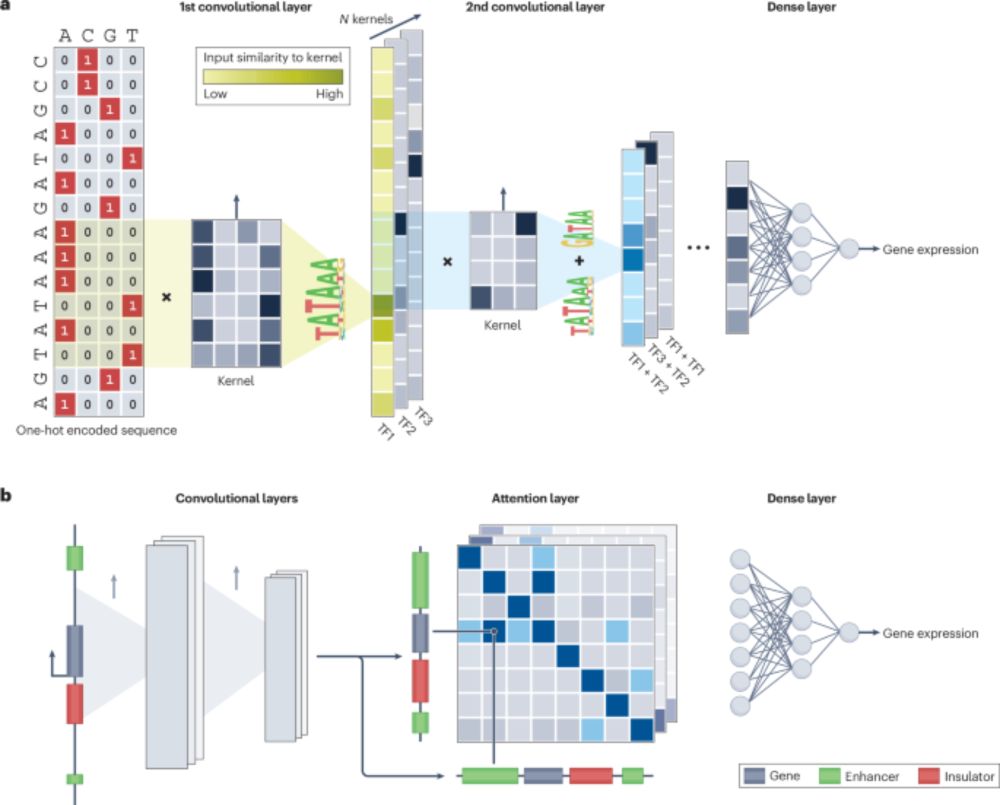
🐕scooby: Modeling multi-modal genomic profiles from DNA sequence at single-cell resolution
📅Wed May 7, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky
🐕scooby: Modeling multi-modal genomic profiles from DNA sequence at single-cell resolution
📅Wed May 7, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...



doi.org/10.1101/2024...

doi.org/10.1101/2024...


