Hsiu-Chuan Lin
@hsiuchuanlin.bsky.social
Group leader @crg.eu | Cell fate engineering and single-cell technology | A Taiwanese 🇹🇼
Pinned
🎉 Two @erc.europa.eu Starting Grants awarded to CRG researchers! DECODE-PMD @markushopfler.bsky.social explores how half-made proteins can trigger deletion of their own mRNA. DECIPHER @hsiuchuanlin.bsky.social decodes gene networks to program precise, mature human brain cells #ERCStG
Read more 👇👇👇
Read more 👇👇👇

Thrilled to share that our group will be supported by an ERC Starting Grant! 🚀
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
Reposted by Hsiu-Chuan Lin
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...

October 16, 2025 at 6:23 AM
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
Thrilled to share that our group will be supported by an ERC Starting Grant! 🚀
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
🎉 Two @erc.europa.eu Starting Grants awarded to CRG researchers! DECODE-PMD @markushopfler.bsky.social explores how half-made proteins can trigger deletion of their own mRNA. DECIPHER @hsiuchuanlin.bsky.social decodes gene networks to program precise, mature human brain cells #ERCStG
Read more 👇👇👇
Read more 👇👇👇

September 4, 2025 at 1:24 PM
Thrilled to share that our group will be supported by an ERC Starting Grant! 🚀
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
With the project 'DECIPHER', we will decode and perturb gene regulation to engineer human cell subtypes, niches, and maturation.
We’ll soon be recruiting at all levels — stay tuned for opportunities to join us!
Reposted by Hsiu-Chuan Lin
Thrilled to share 🎉 I’m starting my lab at University of Zurich,
DMLS as Assistant Professor (tenure track) from Jan 2026!
The Neural MorphoGenomics & Developmental Dynamics Lab will be exploring how genes + morphogenesis shape brain development with organoids, imaging & spatial genomics 🧠🔬🧬
DMLS as Assistant Professor (tenure track) from Jan 2026!
The Neural MorphoGenomics & Developmental Dynamics Lab will be exploring how genes + morphogenesis shape brain development with organoids, imaging & spatial genomics 🧠🔬🧬




September 3, 2025 at 10:43 PM
Thrilled to share 🎉 I’m starting my lab at University of Zurich,
DMLS as Assistant Professor (tenure track) from Jan 2026!
The Neural MorphoGenomics & Developmental Dynamics Lab will be exploring how genes + morphogenesis shape brain development with organoids, imaging & spatial genomics 🧠🔬🧬
DMLS as Assistant Professor (tenure track) from Jan 2026!
The Neural MorphoGenomics & Developmental Dynamics Lab will be exploring how genes + morphogenesis shape brain development with organoids, imaging & spatial genomics 🧠🔬🧬
Reposted by Hsiu-Chuan Lin
Final version @science.org "Recent evolution of the developing human intestine affects metabolic and barrier functions". Evolutionary preparation for microbiota, diet, and pathogen exposure is fascinating! Thank you @erc.europa.eu for supporting our vision!
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

August 22, 2025 at 9:04 PM
Final version @science.org "Recent evolution of the developing human intestine affects metabolic and barrier functions". Evolutionary preparation for microbiota, diet, and pathogen exposure is fascinating! Thank you @erc.europa.eu for supporting our vision!
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Reposted by Hsiu-Chuan Lin
Please RP.
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...
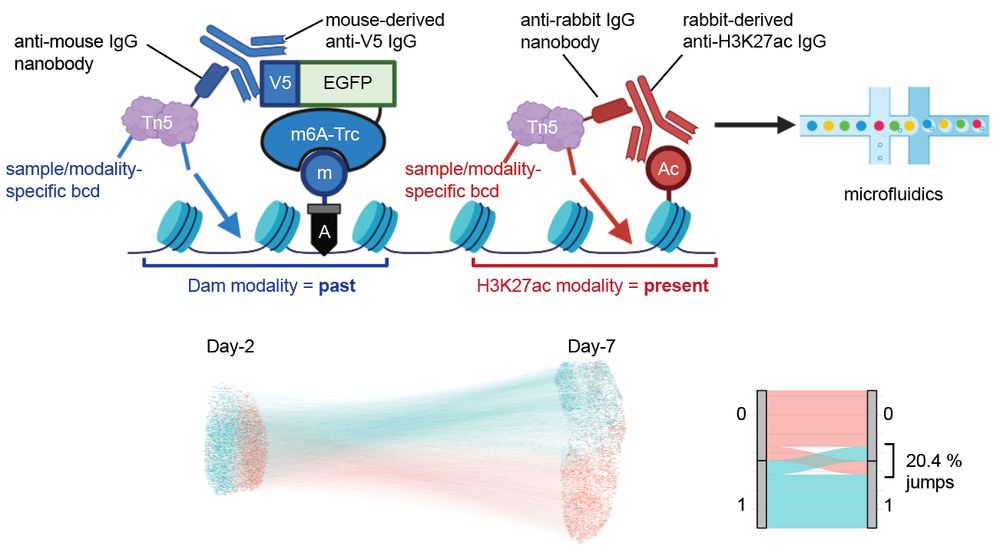
August 16, 2025 at 7:15 AM
Please RP.
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...
Reposted by Hsiu-Chuan Lin
To all post-docs: The Genome Biology dept @embl.org
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

Group Leader - Genome Biology Unit
Are you ready to lead groundbreaking research in Genome Biology? Join us at EMBL! We are seeking a motivated scientist to lead an independent research group addressing exciting and original biological...
embl.wd103.myworkdayjobs.com
July 30, 2025 at 1:41 PM
To all post-docs: The Genome Biology dept @embl.org
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
Reposted by Hsiu-Chuan Lin
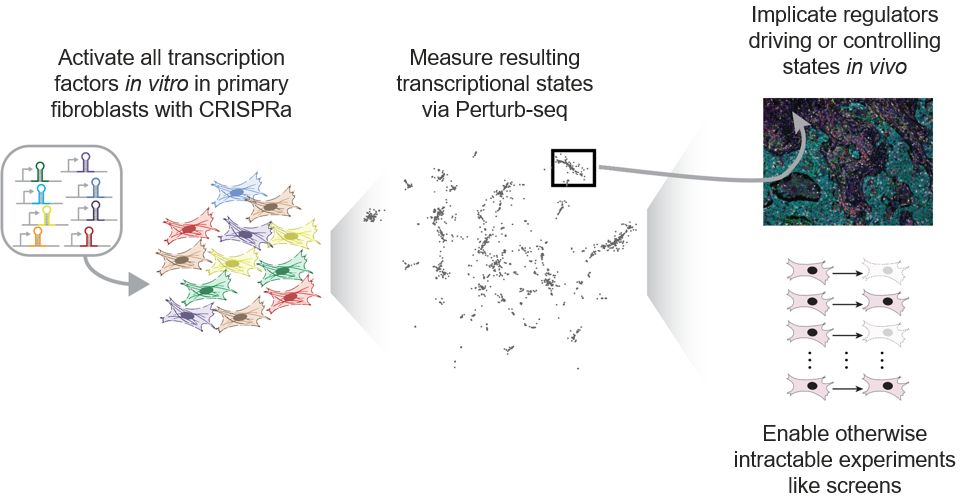
Our paper is now out in final form at Nature Genetics! For those who missed the preprint, we used large-scale Perturb-seq targeting transcription factors to push primary fibroblasts into diverse transcriptional states, including those observed in cell atlas studies.
August 6, 2025 at 3:14 PM
Our paper is now out in final form at Nature Genetics! For those who missed the preprint, we used large-scale Perturb-seq targeting transcription factors to push primary fibroblasts into diverse transcriptional states, including those observed in cell atlas studies.
Reposted by Hsiu-Chuan Lin
🧬🧬🧬 New review from the lab:
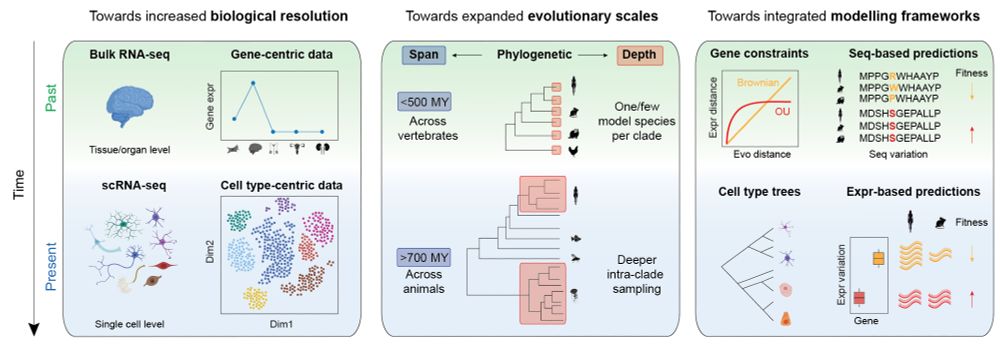
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
August 6, 2025 at 9:16 AM
🧬🧬🧬 New review from the lab:
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
Reposted by Hsiu-Chuan Lin
Just out in @science.org
Silencing mitochondrial gene expression in living cells | Science www.science.org/doi/10.1126/...
Silencing mitochondrial gene expression in living cells | Science www.science.org/doi/10.1126/...
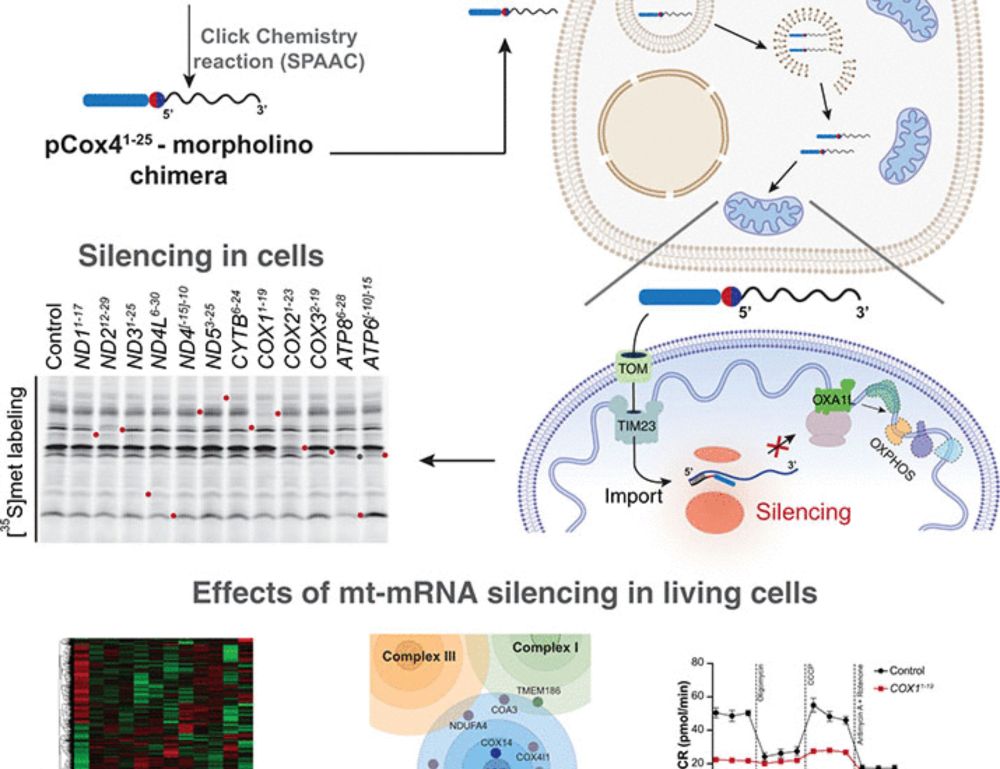
Silencing mitochondrial gene expression in living cells
Mitochondria fulfill central functions in metabolism and energy supply. They express their own genome, which encodes key subunits of the oxidative phosphorylation system. However, the central mechanis...
www.science.org
August 1, 2025 at 10:36 AM
Just out in @science.org
Silencing mitochondrial gene expression in living cells | Science www.science.org/doi/10.1126/...
Silencing mitochondrial gene expression in living cells | Science www.science.org/doi/10.1126/...
Reposted by Hsiu-Chuan Lin
🎲 Our paper on the genetics, energetics, and allostery in proteins with randomized cores and surfaces is out today @science.org!
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
July 25, 2025 at 6:27 AM
🎲 Our paper on the genetics, energetics, and allostery in proteins with randomized cores and surfaces is out today @science.org!
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
Reposted by Hsiu-Chuan Lin
Our work bridging enhancer-promoter proximity to phenotypic outcomes in vivo is out! Shout out to @olimpiabompadre.bsky.social, to Marie Kmita's lab, and to all the co-authors. www.nature.com/articles/s41...
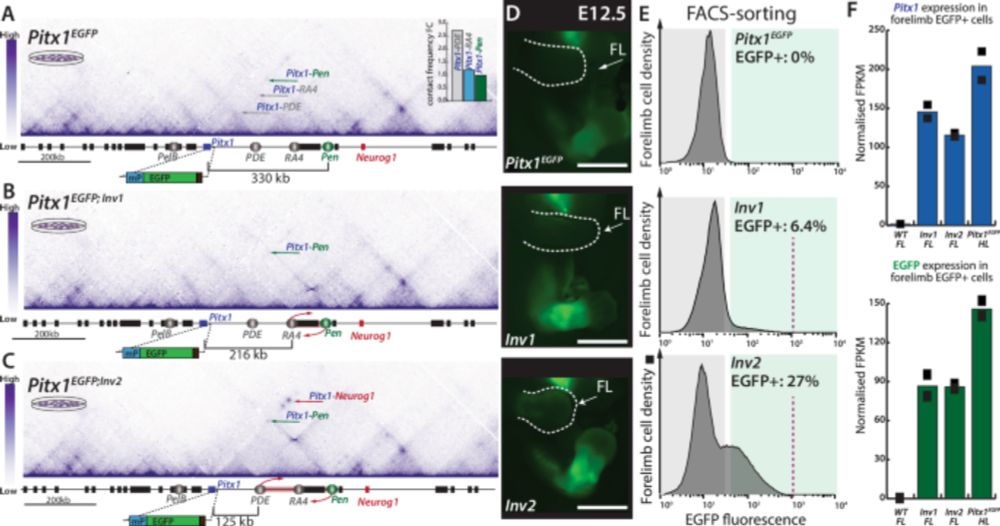
Liebenberg syndrome severity arises from variations in Pitx1 locus topology and proportion of ectopically transcribing cells - Nature Communications
Here the authors show that reducing enhancer-promoter distance at the Pitx1 locus increases proportion of Pitx1 forelimb expressing cells, worsening skeletal defects in Liebenberg syndrome. They also ...
www.nature.com
July 9, 2025 at 11:58 AM
Our work bridging enhancer-promoter proximity to phenotypic outcomes in vivo is out! Shout out to @olimpiabompadre.bsky.social, to Marie Kmita's lab, and to all the co-authors. www.nature.com/articles/s41...
My postdoc work at Treutlein lab and @graycamplab.bsky.social with @jasperjanssens.bsky.social is out in @science.org ! We screen for neuron subtypes using pro-neural TFs + morphogen combinations + scRNA-seq and profiled over 700,000 cells in 480 conditions. www.science.org/doi/10.1126/...
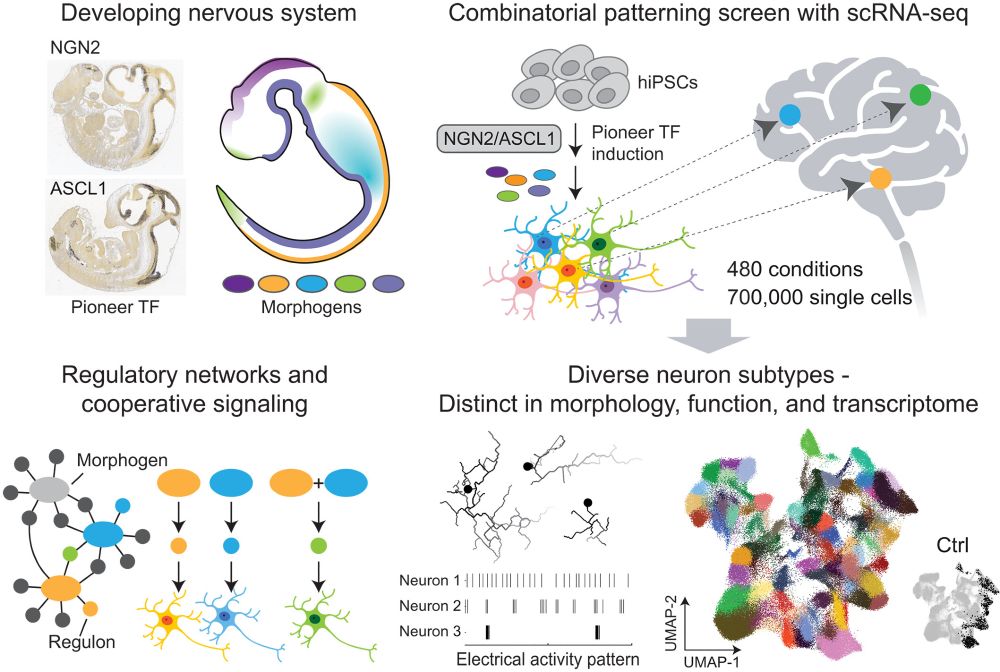
July 12, 2025 at 10:11 PM
My postdoc work at Treutlein lab and @graycamplab.bsky.social with @jasperjanssens.bsky.social is out in @science.org ! We screen for neuron subtypes using pro-neural TFs + morphogen combinations + scRNA-seq and profiled over 700,000 cells in 480 conditions. www.science.org/doi/10.1126/...
Reposted by Hsiu-Chuan Lin
Neuron programming! Pro-neural TFs + 480 morphogen conditions + scRNA-seq --> Diverse iN subtypes of forebrain, midbrain, hindbrain, spinal cord, and PNS. @hsiuchuanlin.bsky.social @jasperjanssens.bsky.social and Treutlein Lab! @science.org www.science.org/doi/10.1126/... #NGN2 #ASCL1
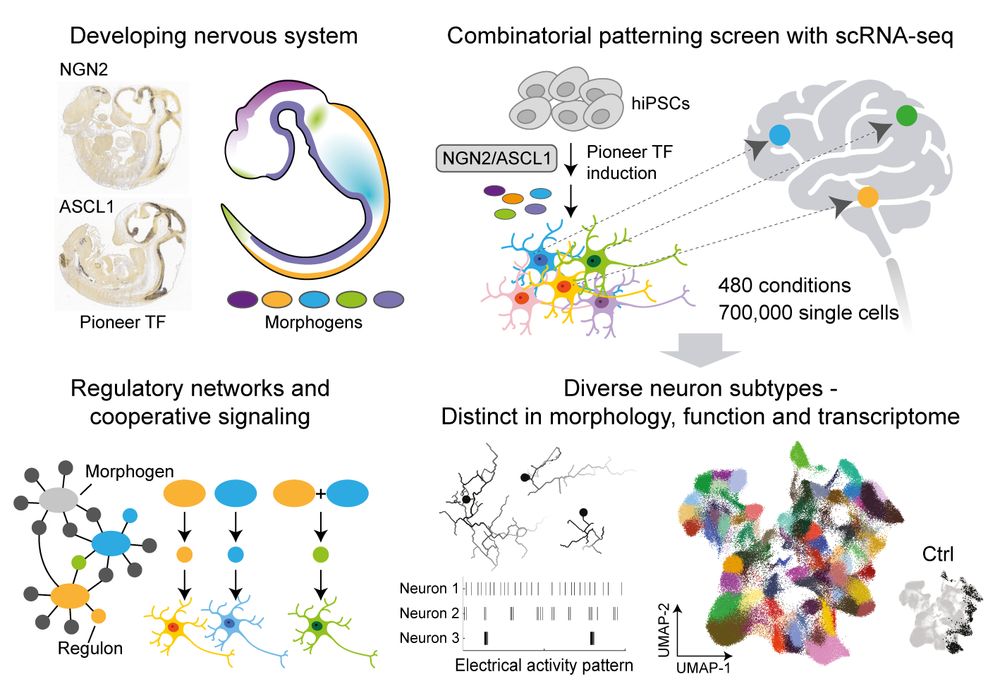
July 11, 2025 at 8:59 PM
Neuron programming! Pro-neural TFs + 480 morphogen conditions + scRNA-seq --> Diverse iN subtypes of forebrain, midbrain, hindbrain, spinal cord, and PNS. @hsiuchuanlin.bsky.social @jasperjanssens.bsky.social and Treutlein Lab! @science.org www.science.org/doi/10.1126/... #NGN2 #ASCL1
Excited to be back in Basel to share my work with old and new friends!
New PI highlight talk! By @hsiuchuanlin.bsky.social who just started her group @crg.eu Barcelona. “Profiling and programming in vitro neuronal diversity at single-cell resolution”

June 24, 2025 at 8:56 PM
Excited to be back in Basel to share my work with old and new friends!
Reposted by Hsiu-Chuan Lin
Just published, expansion in situ genome sequencing, where you can sequence DNA while still inside the cell, mapping its organization relative to proteins and other markers, with the help of expansion microscopy! Led by @jbuenrostro.bksy.social. www.science.org/doi/10.1126/...
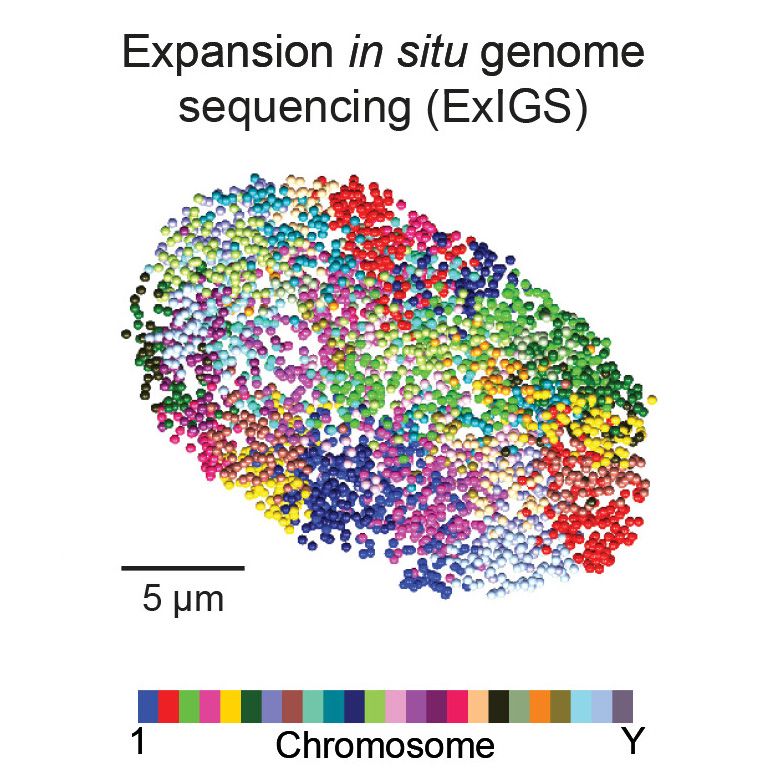
May 30, 2025 at 11:37 AM
Just published, expansion in situ genome sequencing, where you can sequence DNA while still inside the cell, mapping its organization relative to proteins and other markers, with the help of expansion microscopy! Led by @jbuenrostro.bksy.social. www.science.org/doi/10.1126/...
Reposted by Hsiu-Chuan Lin
Our latest "Dynamic Landscape Analysis of Cell Fate Decisions: Predictive Models of Neural Development From Single-Cell Data"
A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...
A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...

Dynamic Landscape Analysis of Cell Fate Decisions: Predictive Models of Neural Development From Single-Cell Data
Building a mechanistic understanding of cell fate decisions remains a fundamental goal of developmental biology, with implications for stem cell therapies, regenerative medicine and understanding dise...
www.biorxiv.org
May 29, 2025 at 12:08 PM
Our latest "Dynamic Landscape Analysis of Cell Fate Decisions: Predictive Models of Neural Development From Single-Cell Data"
A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...
A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...
Reposted by Hsiu-Chuan Lin
Out @nature.com: Clonal tracing with somatic epimutations
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
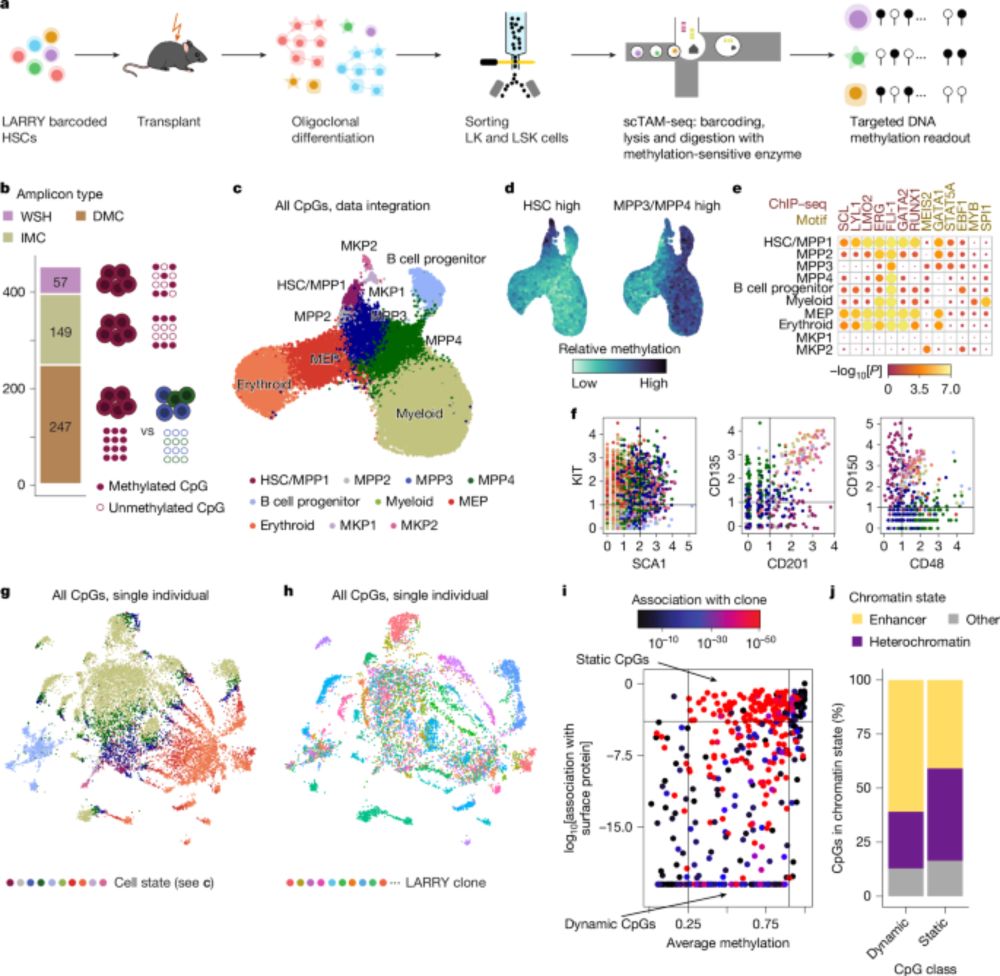
Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
May 21, 2025 at 3:44 PM
Out @nature.com: Clonal tracing with somatic epimutations
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
Reposted by Hsiu-Chuan Lin
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
www.cell.com
May 8, 2025 at 4:07 PM
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
Reposted by Hsiu-Chuan Lin
Registration for Single-cell genomics 2025 is now open! This year we’re back in beautiful Stockholm archipelago. We have a special session on clinical applications, but we also invite abstracts from both PIs and trainees, with plenty of slots for selected talks. conferences.weizmann.ac.il/SCG2025/
Single cell genomics 2025 | SCG 2025
conferences.weizmann.ac.il
April 28, 2025 at 1:01 PM
Registration for Single-cell genomics 2025 is now open! This year we’re back in beautiful Stockholm archipelago. We have a special session on clinical applications, but we also invite abstracts from both PIs and trainees, with plenty of slots for selected talks. conferences.weizmann.ac.il/SCG2025/
Reposted by Hsiu-Chuan Lin
Happy to share our resource for blood vessel organoids!
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
Now online! Fate and state transitions during human blood vessel organoid development

Fate and state transitions during human blood vessel organoid development
Single-cell multiomic reconstruction of the human blood vessel organoid system allows for classifying and modeling disease and cell-type-specific states, demonstrating its potential for translational research.
dlvr.it
April 21, 2025 at 6:39 PM
Happy to share our resource for blood vessel organoids!
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
Reposted by Hsiu-Chuan Lin
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db

CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
April 18, 2025 at 10:19 AM
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db
Reposted by Hsiu-Chuan Lin
New tool 💪 from @steinaerts.bsky.social lab: CREsted, a DL-based approach for predicting cell type-specific reg. elements, applying it to many datasets, incl. mouse cortex, human PBMCs & glioblastoma, demonstrating its ability to identify key TF binding sites: www.biorxiv.org/content/10.1...

CREsted: modeling genomic and synthetic cell type-specific enhancers across tissues and species
Sequence-based deep learning models have become the state of the art for the analysis of the genomic regulatory code. Particularly for transcriptional enhancers, deep learning models excel at decipher...
www.biorxiv.org
April 6, 2025 at 6:10 PM
New tool 💪 from @steinaerts.bsky.social lab: CREsted, a DL-based approach for predicting cell type-specific reg. elements, applying it to many datasets, incl. mouse cortex, human PBMCs & glioblastoma, demonstrating its ability to identify key TF binding sites: www.biorxiv.org/content/10.1...
A fun collaboration with @nazbukina.bsky.social @zhisonghe.bsky.social at @Treutleinlab @graycamplab.bsky.social for our posterior brain organoid atlas and morphogen screens!
Excited to share our latest preprint, presenting a multi-omic human neural organoid cell atlas of the posterior brain! 🧠🔬
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!

March 24, 2025 at 3:15 PM
A fun collaboration with @nazbukina.bsky.social @zhisonghe.bsky.social at @Treutleinlab @graycamplab.bsky.social for our posterior brain organoid atlas and morphogen screens!
Reposted by Hsiu-Chuan Lin
Awesome summary of the field. An important point is to separate the design method from the oracle model being used. Sometimes, people say they're proposing a new design method but mean a cool new oracle model.
Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...
Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...

Modelling and design of transcriptional enhancers - Nature Reviews Bioengineering
Enhancers are genomic elements critical for regulating gene expression. In this Review, the authors discuss how sequence-to-function models can be used to unravel the rules underlying enhancer activit...
www.nature.com
March 3, 2025 at 6:58 PM
Awesome summary of the field. An important point is to separate the design method from the oracle model being used. Sometimes, people say they're proposing a new design method but mean a cool new oracle model.
Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...
Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...

