
jonas
@josch1.bsky.social
dev bio. ML. organoids. single cells @IHB
Pinned
jonas
@josch1.bsky.social
· Apr 18

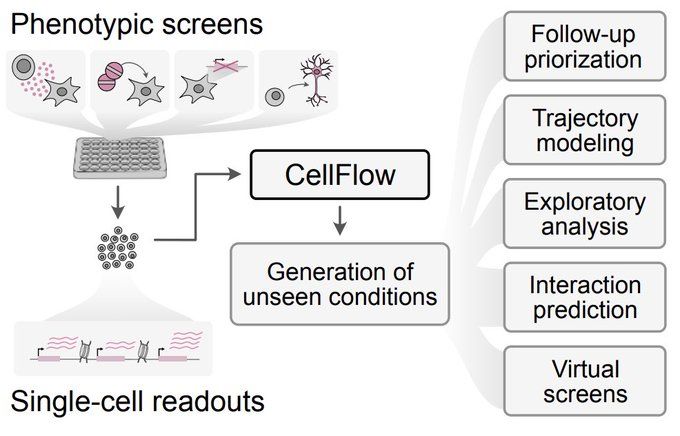
CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db
Yeah, actually, why?
New blog post: Why do we still publish in scientific journals ? What is holding us back from letting go of traditional publishing? No major insight here but I am curious what others think. www.evocellnet.com/2025/07/why-...

Why do we still publish in scientific journals ?
We publish in scientific journals to disclose our discoveries, such that others can build upon them. But we now have preprint servers and we...
www.evocellnet.com
July 23, 2025 at 1:41 PM
Yeah, actually, why?
Reposted by jonas
Yes yes I know. Most science has been hypey forever. But it's hyperaccelated now with this weird meld of academic science & startup culture + social media and highly skilled amplification by marketing & PR teams. 3/
July 10, 2025 at 11:20 PM
Yes yes I know. Most science has been hypey forever. But it's hyperaccelated now with this weird meld of academic science & startup culture + social media and highly skilled amplification by marketing & PR teams. 3/
Reposted by jonas
One thing that really bothers me with the new "virtual cell" terminology is that it is currently largely focused on a very narrow definition of models that can predict effects of trans perturbations (gene dosage, drugs etc) on gene expression. 1/
June 28, 2025 at 10:38 AM
One thing that really bothers me with the new "virtual cell" terminology is that it is currently largely focused on a very narrow definition of models that can predict effects of trans perturbations (gene dosage, drugs etc) on gene expression. 1/
Reposted by jonas
Thrilled to share our Human Endoderm-derived Organoid Cell Atlas (HEOCA). We compare organoids with primary tissues, assess culture strategies, and explore disease models. In collaboration with Lennard Halle, supervised by
@graycamplab.bsky.social, @fabiantheis.bsky.social & @TreutleinLab. @IHB.
@graycamplab.bsky.social, @fabiantheis.bsky.social & @TreutleinLab. @IHB.
Integrated Organoid Atlas benchmarks new protocols, even more impressive as a Highly Variable Cohort for comparison against perturbation and disease models: nature.com/articles/s41... Lead by @quanxu.bsky.social, in collaboration with Lennard Halle and @fabiantheis.bsky.social

An integrated transcriptomic cell atlas of human endoderm-derived organoids - Nature Genetics
The human endoderm-derived organoid cell atlas (HEOCA) presents an integrative analysis of single-cell transcriptomes across different conditions, sources and protocols. It compares cell types and sta...
nature.com
May 13, 2025 at 4:51 AM
Thrilled to share our Human Endoderm-derived Organoid Cell Atlas (HEOCA). We compare organoids with primary tissues, assess culture strategies, and explore disease models. In collaboration with Lennard Halle, supervised by
@graycamplab.bsky.social, @fabiantheis.bsky.social & @TreutleinLab. @IHB.
@graycamplab.bsky.social, @fabiantheis.bsky.social & @TreutleinLab. @IHB.
Reposted by jonas
Modular integration of epithelial, microbial, and immune components to study human intestinal biology. Amazing team project with Rubén López-Sandoval, Marius F. Harter, Qianhui Yu, Matthias Lütolf, Mikhail Nikolaev, and Nikolche Gjorevski! biorxiv.org/content/10.1...
May 9, 2025 at 12:54 PM
Modular integration of epithelial, microbial, and immune components to study human intestinal biology. Amazing team project with Rubén López-Sandoval, Marius F. Harter, Qianhui Yu, Matthias Lütolf, Mikhail Nikolaev, and Nikolche Gjorevski! biorxiv.org/content/10.1...
Reposted by jonas
The first two integrated HCA Organoid Cell Atlases, from brain and endoderm tissues, are available on the HCA Data Portal. Congratulations to all involved! 🖥️ 🧬 data.humancellatlas.org/hca-bio-netw...

April 28, 2025 at 4:21 PM
The first two integrated HCA Organoid Cell Atlases, from brain and endoderm tissues, are available on the HCA Data Portal. Congratulations to all involved! 🖥️ 🧬 data.humancellatlas.org/hca-bio-netw...
Reposted by jonas
It was fun to learn about blood vessel organoids together with @marinanikolova.bsky.social and Barbara Treutlein! Thanks to Reiner Wimmer and @penningerlab.bsky.social for introducing us to this system! www.cell.com/cell/fulltex... Great support from editors and reviewers @cellpress.bsky.social!
April 28, 2025 at 2:33 PM
It was fun to learn about blood vessel organoids together with @marinanikolova.bsky.social and Barbara Treutlein! Thanks to Reiner Wimmer and @penningerlab.bsky.social for introducing us to this system! www.cell.com/cell/fulltex... Great support from editors and reviewers @cellpress.bsky.social!
Reposted by jonas
Our new call for VIB.AI group leaders is online! Junior and senior positions available to develop innovative ML methods in biology. With professorship at CS or Medical Faculty. Deadline 14th June. DM for more info.
We are looking for additional Group Leaders to join the growing computational biology community at VIB.
If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906
If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906

April 26, 2025 at 11:00 AM
Our new call for VIB.AI group leaders is online! Junior and senior positions available to develop innovative ML methods in biology. With professorship at CS or Medical Faculty. Deadline 14th June. DM for more info.
This was such a fun project! A while back, Daniil and I played with flow matching for pred. modeling of cell fate engineering and then teamed up with @dominik1klein.bsky.social and an amazing team @fabiantheis.bsky.social, to build a (close to) universal framework for modeling phenotypic screens 🧵
From cell lines to full embryos, drug treatments to genetic perturbations, neuron engineering to virtual organoid screens — odds are there’s something in it for you!
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1
April 24, 2025 at 4:32 PM
This was such a fun project! A while back, Daniil and I played with flow matching for pred. modeling of cell fate engineering and then teamed up with @dominik1klein.bsky.social and an amazing team @fabiantheis.bsky.social, to build a (close to) universal framework for modeling phenotypic screens 🧵
Reposted by jonas
Happy to share our resource for blood vessel organoids!
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
Now online! Fate and state transitions during human blood vessel organoid development

Fate and state transitions during human blood vessel organoid development
Single-cell multiomic reconstruction of the human blood vessel organoid system allows for classifying and modeling disease and cell-type-specific states, demonstrating its potential for translational research.
dlvr.it
April 21, 2025 at 6:39 PM
Happy to share our resource for blood vessel organoids!
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
@zhisonghe.bsky.social, Makiko Seimiya, Gustav Jonsson, Wuji Cao, Ryo Okuda, Reiner Wimmer, Ryoko Okamoto
Barbara Treutlein, @graycamplab.bsky.social, @penningerlab.bsky.social
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db

CellFlow enables generative single-cell phenotype modeling with flow matching
High-content phenotypic screens provide a powerful strategy for studying biological systems, but the scale of possible perturbations and cell states makes exhaustive experiments unfeasible. Computatio...
www.biorxiv.org
April 18, 2025 at 10:19 AM
Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db
Reposted by jonas
Excited to share our latest preprint, presenting a multi-omic human neural organoid cell atlas of the posterior brain! 🧠🔬
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!

March 24, 2025 at 12:42 PM
Excited to share our latest preprint, presenting a multi-omic human neural organoid cell atlas of the posterior brain! 🧠🔬
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!
Reposted by jonas
Good to see moscot-tools.org published in @nature.com ! We made existing Optimal Transport (OT) applications in single-cell genomics scalable and multimodal, added a novel spatiotemporal trajectory inference method and found exciting new biology in the pancreas! tinyurl.com/33zuwsep
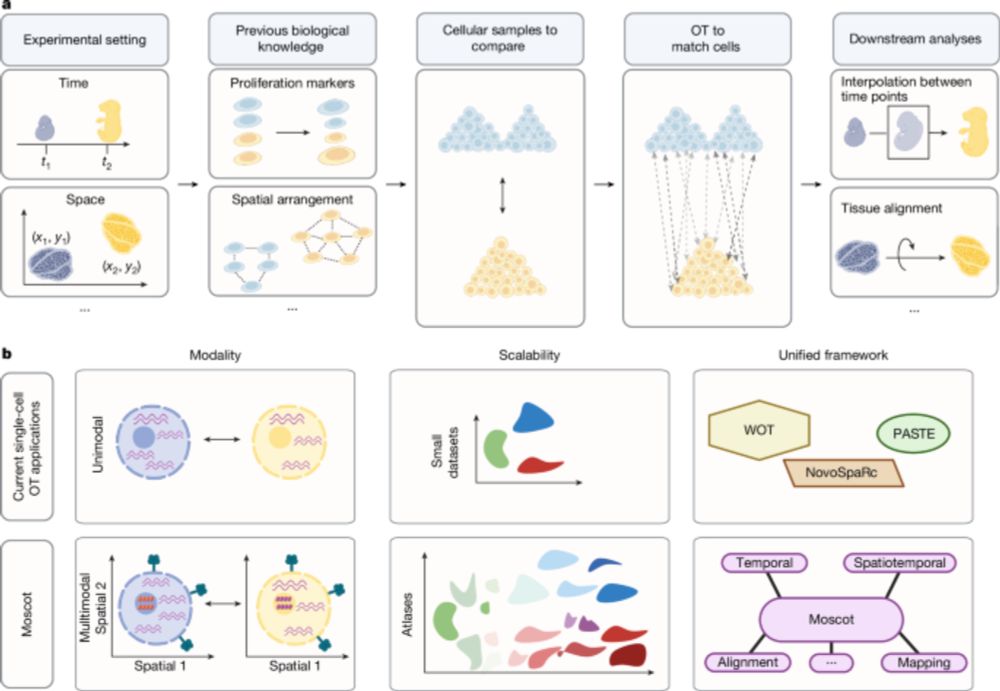
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
tinyurl.com
January 23, 2025 at 8:42 AM
Good to see moscot-tools.org published in @nature.com ! We made existing Optimal Transport (OT) applications in single-cell genomics scalable and multimodal, added a novel spatiotemporal trajectory inference method and found exciting new biology in the pancreas! tinyurl.com/33zuwsep
Reposted by jonas
Excited to see Moscot (moscot-tools.org) published in @Nature! We scaled Optimal Transport (OT) in single-cell genomics & added multimodality together with spatiotemporal trajectory inference, finding exciting new biology in the pancreas! 🚀 Read at www.nature.com/articles/s41...
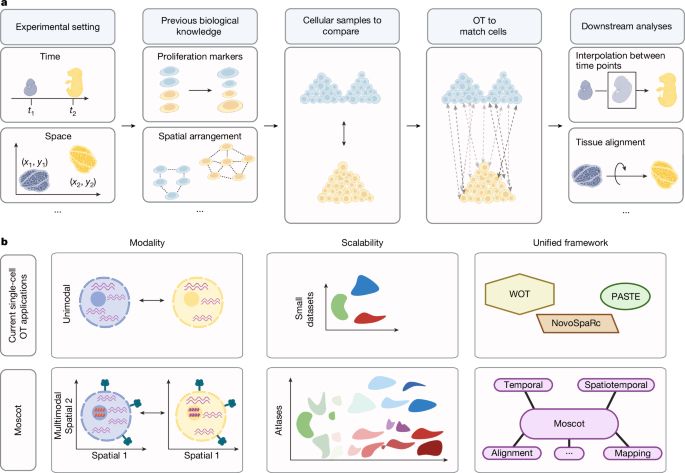
Mapping cells through time and space with moscot - Nature
Moscot is an optimal transport approach that overcomes current limitations of similar methods to enable multimodal, scalable and consistent single-cell analyses of datasets across spatial and temporal...
www.nature.com
January 22, 2025 at 10:46 PM
Excited to see Moscot (moscot-tools.org) published in @Nature! We scaled Optimal Transport (OT) in single-cell genomics & added multimodality together with spatiotemporal trajectory inference, finding exciting new biology in the pancreas! 🚀 Read at www.nature.com/articles/s41...
💯
The only sort of gatekeeper I wish we saw more of were those who wanted better software. Who cares if your model performs the best if it's unusable, or if your pipeline aligs reads faster if it only works on your local work computer? Or if there's no documentation on how to use any of it.
January 6, 2025 at 8:47 PM
💯
Reposted by jonas
I’m always looking for things that explain a lot but that people have a hard time remembering.
Examples: Air is stuff. Pee comes from blood. All land vertebrates have a single common ancestor. Venus is bright enough to cast visible shadows. Clouds are heavy. Bones are alive.
Any others you know?
Examples: Air is stuff. Pee comes from blood. All land vertebrates have a single common ancestor. Venus is bright enough to cast visible shadows. Clouds are heavy. Bones are alive.
Any others you know?
December 28, 2024 at 8:06 PM
I’m always looking for things that explain a lot but that people have a hard time remembering.
Examples: Air is stuff. Pee comes from blood. All land vertebrates have a single common ancestor. Venus is bright enough to cast visible shadows. Clouds are heavy. Bones are alive.
Any others you know?
Examples: Air is stuff. Pee comes from blood. All land vertebrates have a single common ancestor. Venus is bright enough to cast visible shadows. Clouds are heavy. Bones are alive.
Any others you know?
Reposted by jonas
If diving into scientific papers isn’t on your pre-Christmas to-do list, @spiegel.de has you covered! 📰 They featured our Neural Organoid Atlas in a piece on the Human Cell Atlas and related advancements in machine learning. 🤖🧬 It’s a great overview of this exciting field and the work behind it.

Meinung: »Human Cell Atlas«: Das wird die wichtigste Wissenschaft des 21. Jahrhunderts - Kolumne
Gerade ist ein ganzes Bündel Publikationen aus einem einzigen Forschungsprojekt erschienen. Sie weisen in die Zukunft einer neuen Wissenschaft: Lernende Maschinen helfen jetzt, die Maschinerie des Leb...
www.spiegel.de
December 19, 2024 at 4:05 PM
If diving into scientific papers isn’t on your pre-Christmas to-do list, @spiegel.de has you covered! 📰 They featured our Neural Organoid Atlas in a piece on the Human Cell Atlas and related advancements in machine learning. 🤖🧬 It’s a great overview of this exciting field and the work behind it.
Bit late with this, but super happy that our neural organoid atlas is out in Nature! Really fun collab with @zhisonghe.bsky.social and @le-and-er.bsky.social between the Treutlein lab, @fabiantheis.bsky.social and @graycamplab.bsky.social.
www.nature.com/articles/s41...
www.nature.com/articles/s41...

An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com
December 13, 2024 at 2:27 PM
Bit late with this, but super happy that our neural organoid atlas is out in Nature! Really fun collab with @zhisonghe.bsky.social and @le-and-er.bsky.social between the Treutlein lab, @fabiantheis.bsky.social and @graycamplab.bsky.social.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by jonas
📢 There are 2 open PhD positions in my group at CRG Barcelona! We are looking for candidates interested in cell fate engineering, gene regulation, and single-cell technologies with experience in both wet/dry labs.
Please help spread the word 🙌
Apply here ➡️ www.crg.eu/en/content/t...
Please help spread the word 🙌
Apply here ➡️ www.crg.eu/en/content/t...
Centre for Genomic Regulation Website
www.crg.eu
November 28, 2024 at 11:52 PM
📢 There are 2 open PhD positions in my group at CRG Barcelona! We are looking for candidates interested in cell fate engineering, gene regulation, and single-cell technologies with experience in both wet/dry labs.
Please help spread the word 🙌
Apply here ➡️ www.crg.eu/en/content/t...
Please help spread the word 🙌
Apply here ➡️ www.crg.eu/en/content/t...
Reposted by jonas
Glad that our neural organoid atlas is out @Nature! Many interesting analyses for in vitro-in vivo comparisons beyond neural organoid setting - here eg fig 4e, where we visualize on primary reference which brain regions are not yet covered by organoids. Exciting for exp design!

November 22, 2024 at 5:25 PM
Glad that our neural organoid atlas is out @Nature! Many interesting analyses for in vitro-in vivo comparisons beyond neural organoid setting - here eg fig 4e, where we visualize on primary reference which brain regions are not yet covered by organoids. Exciting for exp design!
Reposted by jonas
Now in Nature! As a highly collaborative team with people from the Treutlein lab, @fabiantheis.bsky.social and @graycamplab.bsky.social, led by me, @le-and-er.bsky.social and @josch1.bsky.social, we generated the so far largest cell atlas of neural #organoid. www.nature.com/articles/s41...

An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com
November 21, 2024 at 10:20 PM
Now in Nature! As a highly collaborative team with people from the Treutlein lab, @fabiantheis.bsky.social and @graycamplab.bsky.social, led by me, @le-and-er.bsky.social and @josch1.bsky.social, we generated the so far largest cell atlas of neural #organoid. www.nature.com/articles/s41...
Reposted by jonas
Super excited to share our Human Neural Organoid Atlas, now out in Nature! Led by @zhisonghe.bsky.social @josch1.bsky.social, and myself, this resource was created from 36 scRNA-seq datasets—totalling over 1.7 million cells! 🔬✨
www.nature.com/articles/s41...
Find out how it can serve you ⏬
🧵1/8
Find out how it can serve you ⏬
🧵1/8
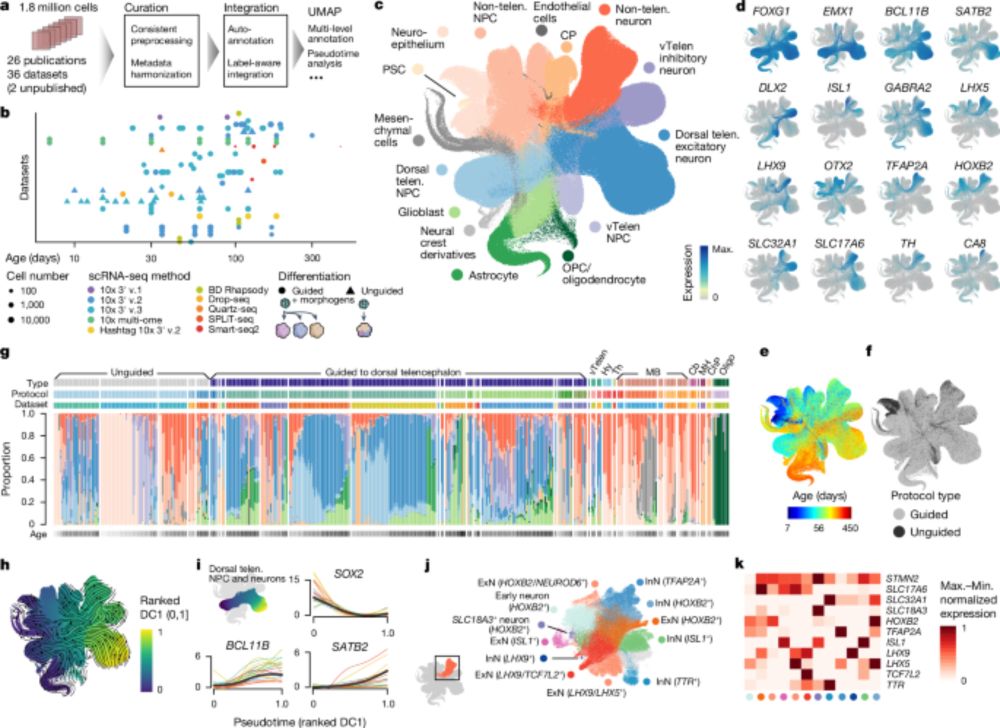
An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com
November 21, 2024 at 10:11 AM
Super excited to share our Human Neural Organoid Atlas, now out in Nature! Led by @zhisonghe.bsky.social @josch1.bsky.social, and myself, this resource was created from 36 scRNA-seq datasets—totalling over 1.7 million cells! 🔬✨
www.nature.com/articles/s41...
Find out how it can serve you ⏬
🧵1/8
Find out how it can serve you ⏬
🧵1/8
Reposted by jonas
A big day of output for the Human Cell Atlas, a global collaborative project with 100 countries to understand our ~37 trillion cells
A Wikipedia of our cells, a "remarkable achievement"
nature.com/articles/d41...
nature.com/immersive/d4...
A Wikipedia of our cells, a "remarkable achievement"
nature.com/articles/d41...
nature.com/immersive/d4...

November 20, 2024 at 5:04 PM
A big day of output for the Human Cell Atlas, a global collaborative project with 100 countries to understand our ~37 trillion cells
A Wikipedia of our cells, a "remarkable achievement"
nature.com/articles/d41...
nature.com/immersive/d4...
A Wikipedia of our cells, a "remarkable achievement"
nature.com/articles/d41...
nature.com/immersive/d4...
Reposted by jonas
The integrated transcriptomic cell atlas of human neural #organoids, Barbara Treutlein's lab, @fabiantheis.bsky.social @graycamplab.bsky.social @sergiuppasca.bsky.social @giorgiaquadrato.bsky.social and colleagues:
www.nature.com/articles/s41...
www.nature.com/articles/s41...
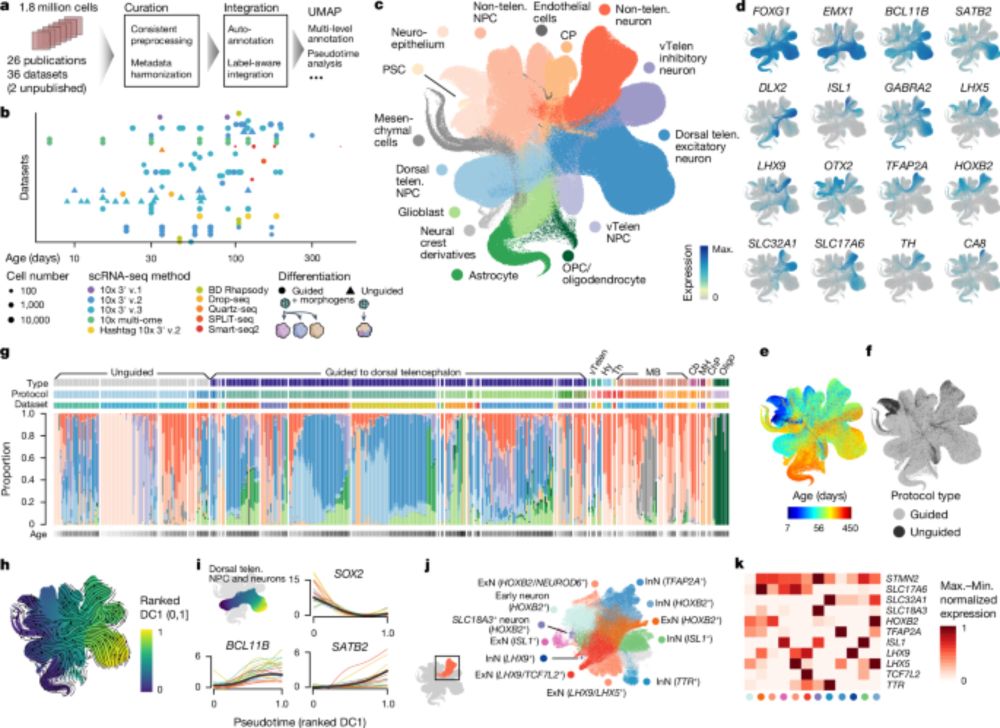
An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com
November 21, 2024 at 6:39 AM
The integrated transcriptomic cell atlas of human neural #organoids, Barbara Treutlein's lab, @fabiantheis.bsky.social @graycamplab.bsky.social @sergiuppasca.bsky.social @giorgiaquadrato.bsky.social and colleagues:
www.nature.com/articles/s41...
www.nature.com/articles/s41...

