Systems Biology of Proteostasis
We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
I’m delighted to share that registration is now open for the UK Proteostasis Meeting 2026, hosted by The Francis Crick Institute on 20–21 July 2026 .
Please register here(lnkd.in/ervXMzWN) and through Eventbrite for payment (lnkd.in/eTxqjnQy)

We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
go.nature.com/4qZM3Cn

go.nature.com/4qZM3Cn
go.nature.com/4qZM3Cn

CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis
www.cell.com/cell/fulltex...
Great collaborative effort - read more from first author @asamelson.bsky.social below:
CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis
www.cell.com/cell/fulltex...
Great collaborative effort - read more from first author @asamelson.bsky.social below:
www.cell.com/molecular-ce...

www.cell.com/molecular-ce...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
We are looking for creative, innovative scientists asking fundamental questions in any area of biology.
Join our vibrant, collaborative, and supportive community!
aprecruit.ucsf.edu/JPF05702

Our new study shows that α-syn fibrils hijack the ESCRT membrane repair system, triggering a feedback loop that worsens aggregation.
You can find it at: authors.elsevier.com/sd/article/S...
Our new study shows that α-syn fibrils hijack the ESCRT membrane repair system, triggering a feedback loop that worsens aggregation.
You can find it at: authors.elsevier.com/sd/article/S...

www.cell.com/cell/fulltex...

www.cell.com/cell/fulltex...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...

#proteostasis #apolipoprotein
rdcu.be/ezRLv

#proteostasis #apolipoprotein
rdcu.be/ezRLv
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
By studying a protein that is difficult to fold, we discover fascinating new mechanisms by which the ribosome supports protein biogenesis.
www.biorxiv.org/content/10.1...
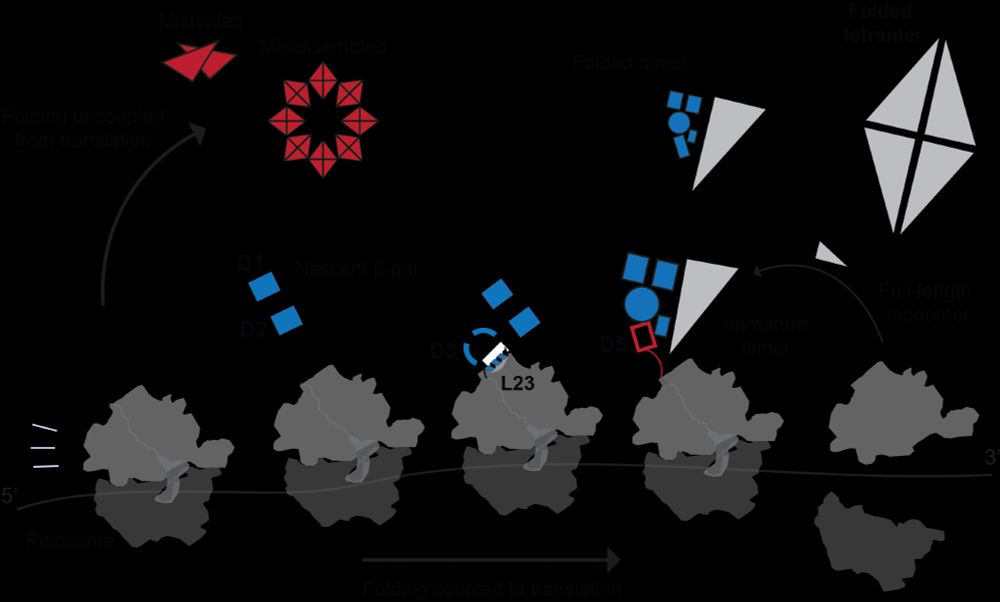
By studying a protein that is difficult to fold, we discover fascinating new mechanisms by which the ribosome supports protein biogenesis.
www.biorxiv.org/content/10.1...
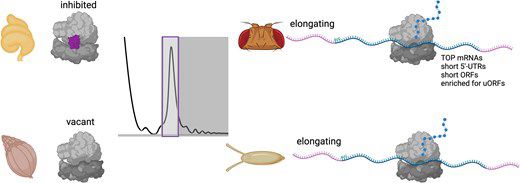
I am super happy to share this review as my first post on Bluesky. After submitting my PhD thesis at the start of the year, my PI Elke Deuerling suggested to write this review with her, covering many of the findings of my thesis. I am excited to see it published today!

I am super happy to share this review as my first post on Bluesky. After submitting my PhD thesis at the start of the year, my PI Elke Deuerling suggested to write this review with her, covering many of the findings of my thesis. I am excited to see it published today!






