The @dfg.de just announced that ImmunoSensation3, focussing on Immune Diversity will be funded!
Let us #celebrate 7 more years of #immunology in #Bonn 🎉
www.immunosensation.de/news/immunos...

The @dfg.de just announced that ImmunoSensation3, focussing on Immune Diversity will be funded!
Let us #celebrate 7 more years of #immunology in #Bonn 🎉
www.immunosensation.de/news/immunos...
© G. Hübl, V. Lannert, M. Thürbach/ECONtribute, J. F. Saba/UKB, S. Wegener/ML4Q; N. Wietrich

© G. Hübl, V. Lannert, M. Thürbach/ECONtribute, J. F. Saba/UKB, S. Wegener/ML4Q; N. Wietrich

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
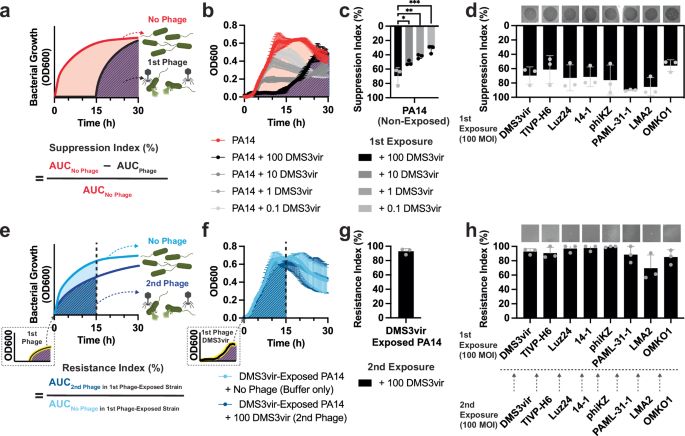
We report a new immune signaling molecule: N7-cADPR. Produced by phage-induced TIRs and activates a defensive bacterial caspase
Congrats Francois Rousset, Ilya Osterman and coauthors!
www.science.org/doi/10.1126/...


We report a new immune signaling molecule: N7-cADPR. Produced by phage-induced TIRs and activates a defensive bacterial caspase
Congrats Francois Rousset, Ilya Osterman and coauthors!
www.science.org/doi/10.1126/...
Way to go Jimbo!

Way to go Jimbo!

Way to go Jimbo!
Thanks to the team including: @philipphendricks.bsky.social @scbinder.bsky.social @gavinhthomas.bsky.social @gha14.bsky.social
Read the full story here:
doi.org/10.1038/s420...

Thanks to the team including: @philipphendricks.bsky.social @scbinder.bsky.social @gavinhthomas.bsky.social @gha14.bsky.social
Read the full story here:
doi.org/10.1038/s420...
Amazing job by @nschneberger.bsky.social and thanks to everyone involved! As always a great team effort!
rdcu.be/d1foN

Amazing job by @nschneberger.bsky.social and thanks to everyone involved! As always a great team effort!
rdcu.be/d1foN
@chrisfrank662.bsky.social , Dominik Schiwietz, Lara Fuss, Hendrik Dietz
www.biorxiv.org/content/10.1...
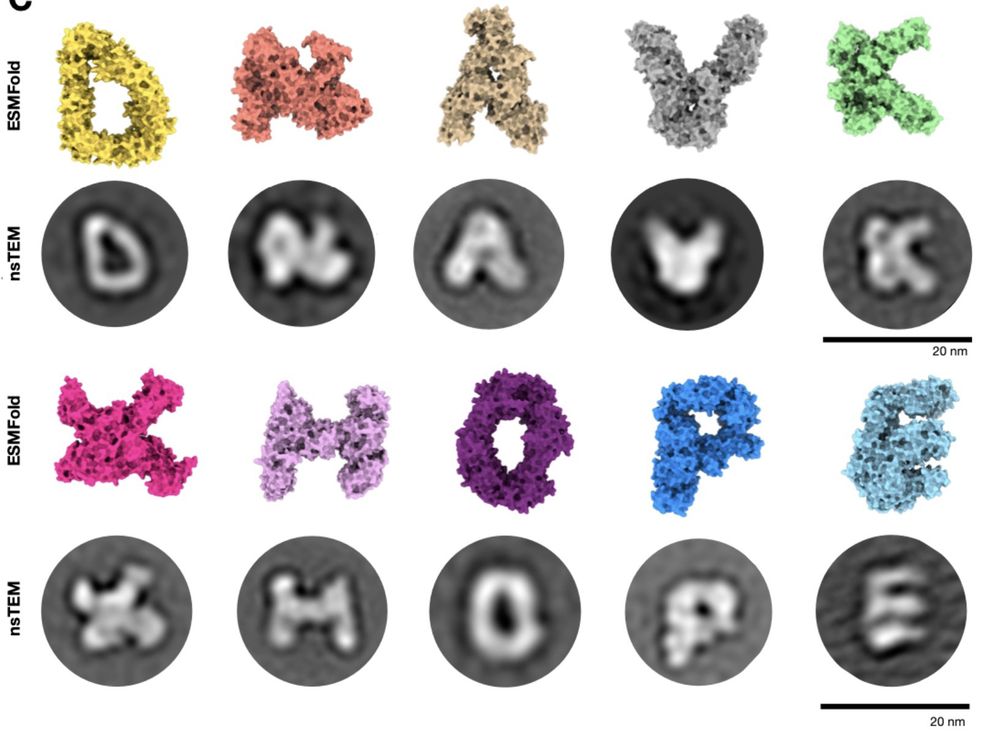
@chrisfrank662.bsky.social , Dominik Schiwietz, Lara Fuss, Hendrik Dietz
www.biorxiv.org/content/10.1...
shorturl.at/88tdH
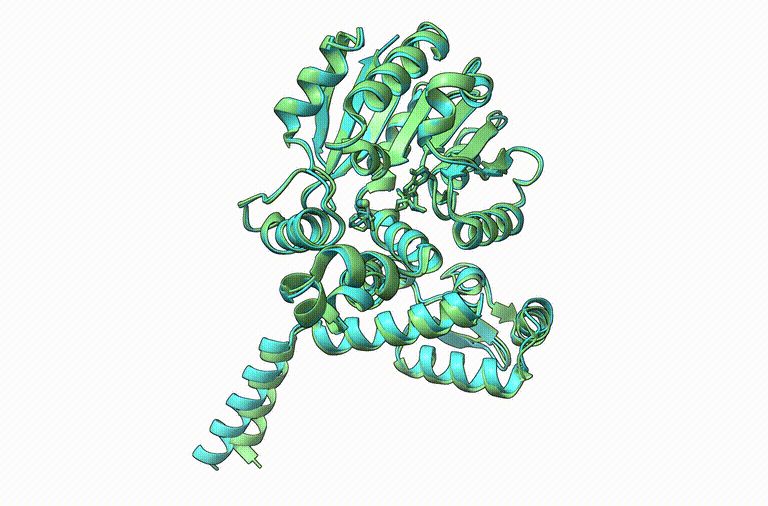
This work was led by SoCoBio DTP student Joe Davies (not on Bluesky yet) and was really fun to work on.
shorturl.at/88tdH
This work was led by SoCoBio DTP student Joe Davies (not on Bluesky yet) and was really fun to work on.


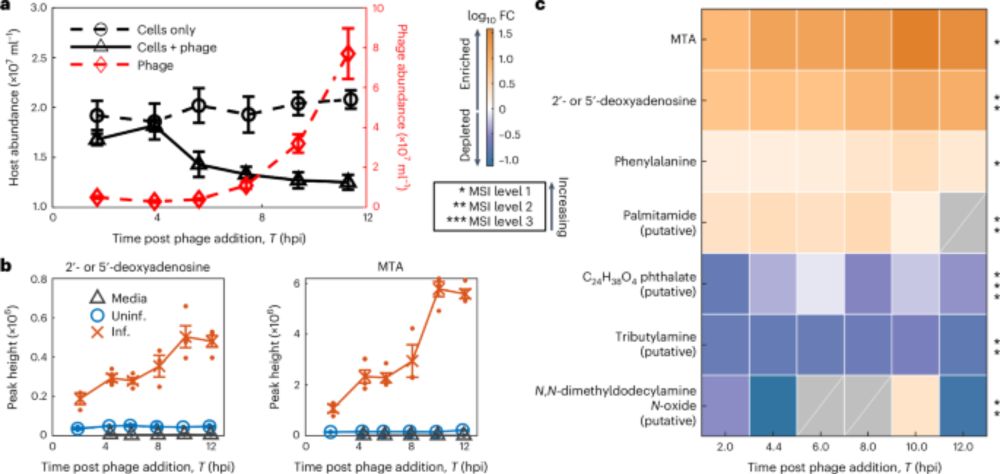
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...