Another reason to be bullish on UK AI. Boltz, a startup that emerged out of MIT to be at the frontier of small molecule and protein design, has moved to London.
I hear cases like this from a few founders, often a mix of fundamentals (talent)+personal.
Another reason to be bullish on UK AI. Boltz, a startup that emerged out of MIT to be at the frontier of small molecule and protein design, has moved to London.
I hear cases like this from a few founders, often a mix of fundamentals (talent)+personal.
I think a PhD is a very special time. You get to challenge yourself, push your boundaries, and grow. My thoughts go against the current AI/academia narrative online, so I hope you find it interesting.
chaitjo.substack.com/p/phd-thesis...

I think a PhD is a very special time. You get to challenge yourself, push your boundaries, and grow. My thoughts go against the current AI/academia narrative online, so I hope you find it interesting.
chaitjo.substack.com/p/phd-thesis...
📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇

📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇
#AISky #AcademicSky

#AISky #AcademicSky


, Felix Faltings, Regina Barzilay, Tommi Jaakkola, and co. We applied BoltzGen to design novel antimicrobial peptides targeting DNA gyrase based on inhibitory protein fragments. (1/n)

, Felix Faltings, Regina Barzilay, Tommi Jaakkola, and co. We applied BoltzGen to design novel antimicrobial peptides targeting DNA gyrase based on inhibitory protein fragments. (1/n)





Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...
Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...

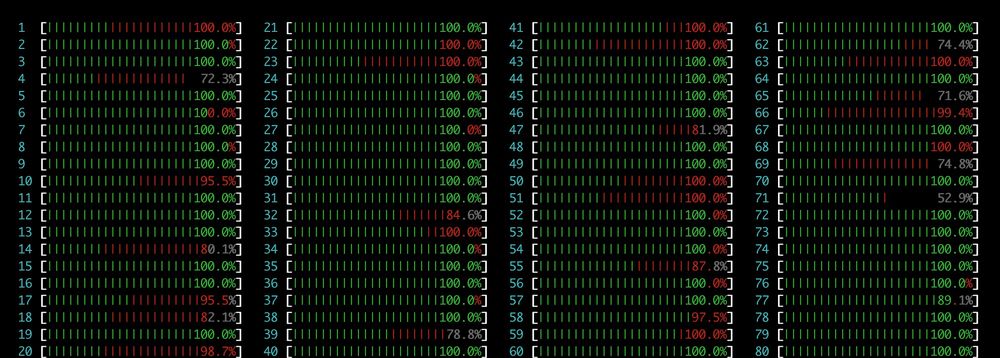

Preprint: doi.org/10.1101/2025...
Preprint: doi.org/10.1101/2025...
youtu.be/iHDauMATkr0?...

youtu.be/iHDauMATkr0?...
- YouTube www.youtube.com/watch?v=LLij...
- Spotify spotifycreators-web.app.link/e/NNmzqnTjgUb
- Apple podcasts.apple.com/us/podcast/h...

- YouTube www.youtube.com/watch?v=LLij...
- Spotify spotifycreators-web.app.link/e/NNmzqnTjgUb
- Apple podcasts.apple.com/us/podcast/h...




Since the release of Boltz-2 last Friday – the new open-source protein structure and protein binding affinity model from MIT & Recursion – we’ve been introducing the model to the broader community and the reception has been terrific. 🧵
Since the release of Boltz-2 last Friday – the new open-source protein structure and protein binding affinity model from MIT & Recursion – we’ve been introducing the model to the broader community and the reception has been terrific. 🧵
👉 Learn more: portal.ml4dd.com #MOML2025
👉 Learn more: portal.ml4dd.com #MOML2025

