
@dgautheret.bsky.social
Trying to make sense of #RNA information.
www.biorxiv.org/cgi/content/...
What’s behind Metapuccino? ☕️, by PhD student Fiona Hak, @camillemrcht.bsky.social and Melina Gallopin. A thread 👇
What’s behind Metapuccino? ☕️, by PhD student Fiona Hak, @camillemrcht.bsky.social and Melina Gallopin. A thread 👇

Metappuccino: Large Language Model-driven Reconstruction of Sequence Read Archive Metadata for Cancer Research
Motivation: High-throughput RNA-sequencing has significantly advanced transcriptomic profiling in oncology. Millions of RNA-seq datasets have accumulated in public databases such as the Sequence Read ...
www.biorxiv.org
November 2, 2025 at 10:14 AM
www.biorxiv.org/cgi/content/...
What’s behind Metapuccino? ☕️, by PhD student Fiona Hak, @camillemrcht.bsky.social and Melina Gallopin. A thread 👇
What’s behind Metapuccino? ☕️, by PhD student Fiona Hak, @camillemrcht.bsky.social and Melina Gallopin. A thread 👇
Reposted
My algorithmic friends (@camillemrcht.bsky.social) doing LLM stuff : www.biorxiv.org/content/10.1...! And also, screaming last names in the author list ;P. Given my level of trust in Camille, though, perhaps it's time for me to engage more seriously with these models in research...

Metappuccino: Large Language Model-driven Reconstruction of Sequence Read Archive Metadata for Cancer Research
Motivation: High-throughput RNA-sequencing has significantly advanced transcriptomic profiling in oncology. Millions of RNA-seq datasets have accumulated in public databases such as the Sequence Read ...
www.biorxiv.org
November 1, 2025 at 3:08 PM
My algorithmic friends (@camillemrcht.bsky.social) doing LLM stuff : www.biorxiv.org/content/10.1...! And also, screaming last names in the author list ;P. Given my level of trust in Camille, though, perhaps it's time for me to engage more seriously with these models in research...
Reposted
Interested in #lncRNA and #ArtificiaIntelligence?
In the frame of our recently founded French-Korean bilateral project DHARP, we are recruiting a post-doc in bioinformatics and artificial intelligence in our team at
@ips2parissaclay.bsky.social
Application limit: 01/12/2025
In the frame of our recently founded French-Korean bilateral project DHARP, we are recruiting a post-doc in bioinformatics and artificial intelligence in our team at
@ips2parissaclay.bsky.social
Application limit: 01/12/2025

October 22, 2025 at 3:27 PM
Interested in #lncRNA and #ArtificiaIntelligence?
In the frame of our recently founded French-Korean bilateral project DHARP, we are recruiting a post-doc in bioinformatics and artificial intelligence in our team at
@ips2parissaclay.bsky.social
Application limit: 01/12/2025
In the frame of our recently founded French-Korean bilateral project DHARP, we are recruiting a post-doc in bioinformatics and artificial intelligence in our team at
@ips2parissaclay.bsky.social
Application limit: 01/12/2025
Reposted
PubMed is running on autopilot during shutdown, but key independent committee has been abolished www.bmj.com/content/391/... 🧪

October 22, 2025 at 5:55 PM
PubMed is running on autopilot during shutdown, but key independent committee has been abolished www.bmj.com/content/391/... 🧪
Reposted
New tool "bwt-svg" for making illustrations of the BWT and the many auxiliary arrays and other structures related to it. Pyodide-based no-installation-necessary interface here: benlangmead.github.io/bwt-svg/. (H/t to @robert.bio for pointing me to pyodide!) Full repo: github.com/benlangmead/....

October 14, 2025 at 8:48 PM
New tool "bwt-svg" for making illustrations of the BWT and the many auxiliary arrays and other structures related to it. Pyodide-based no-installation-necessary interface here: benlangmead.github.io/bwt-svg/. (H/t to @robert.bio for pointing me to pyodide!) Full repo: github.com/benlangmead/....
The MSc. Bioinformatics students of U. Paris-Saclay are organizing the Junior Conference on Computational Biology (JC2B) 2025: AI and predictive models in bioinformatics
November 13, 2025 - I2BC, CNRS, Gif-sur-Yvette, France
Register for free : bioi2.i2bc.paris-saclay.fr/jc2b/#regist...
November 13, 2025 - I2BC, CNRS, Gif-sur-Yvette, France
Register for free : bioi2.i2bc.paris-saclay.fr/jc2b/#regist...

October 1, 2025 at 12:38 PM
The MSc. Bioinformatics students of U. Paris-Saclay are organizing the Junior Conference on Computational Biology (JC2B) 2025: AI and predictive models in bioinformatics
November 13, 2025 - I2BC, CNRS, Gif-sur-Yvette, France
Register for free : bioi2.i2bc.paris-saclay.fr/jc2b/#regist...
November 13, 2025 - I2BC, CNRS, Gif-sur-Yvette, France
Register for free : bioi2.i2bc.paris-saclay.fr/jc2b/#regist...
Reposted
🦠🧍♀️From bacterial to human immunity.
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
www.science.org
July 24, 2025 at 6:23 PM
🦠🧍♀️From bacterial to human immunity.
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
Reposted
Congratulations to Rayan Chiki, (Institut Pasteur) head of the “Sequence Bioinformatics” unit, for securing the ERC Proof of Concept 2025 for his project ENZYMINER! 👏
@rayan.chiki.bsky.social
#Bioinformatics
@rayan.chiki.bsky.social
#Bioinformatics


July 24, 2025 at 3:10 PM
Congratulations to Rayan Chiki, (Institut Pasteur) head of the “Sequence Bioinformatics” unit, for securing the ERC Proof of Concept 2025 for his project ENZYMINER! 👏
@rayan.chiki.bsky.social
#Bioinformatics
@rayan.chiki.bsky.social
#Bioinformatics
Ca a l'air bien, non?
www.nature.com/articles/d41...
www.nature.com/articles/d41...

How to speed up peer review: make applicants mark one another
‘Distributed peer review’ of grants makes process more than twice as fast — and includes some cheat-prevention measures.
www.nature.com
July 23, 2025 at 7:46 AM
Ca a l'air bien, non?
www.nature.com/articles/d41...
www.nature.com/articles/d41...
Reposted
Paper Alert!
Our preprint on the K2R index, being able to efficiently associate kmers to the reads containing them is finally out there!
A thread!
academic.oup.com/bioinformati...
Our preprint on the K2R index, being able to efficiently associate kmers to the reads containing them is finally out there!
A thread!
academic.oup.com/bioinformati...
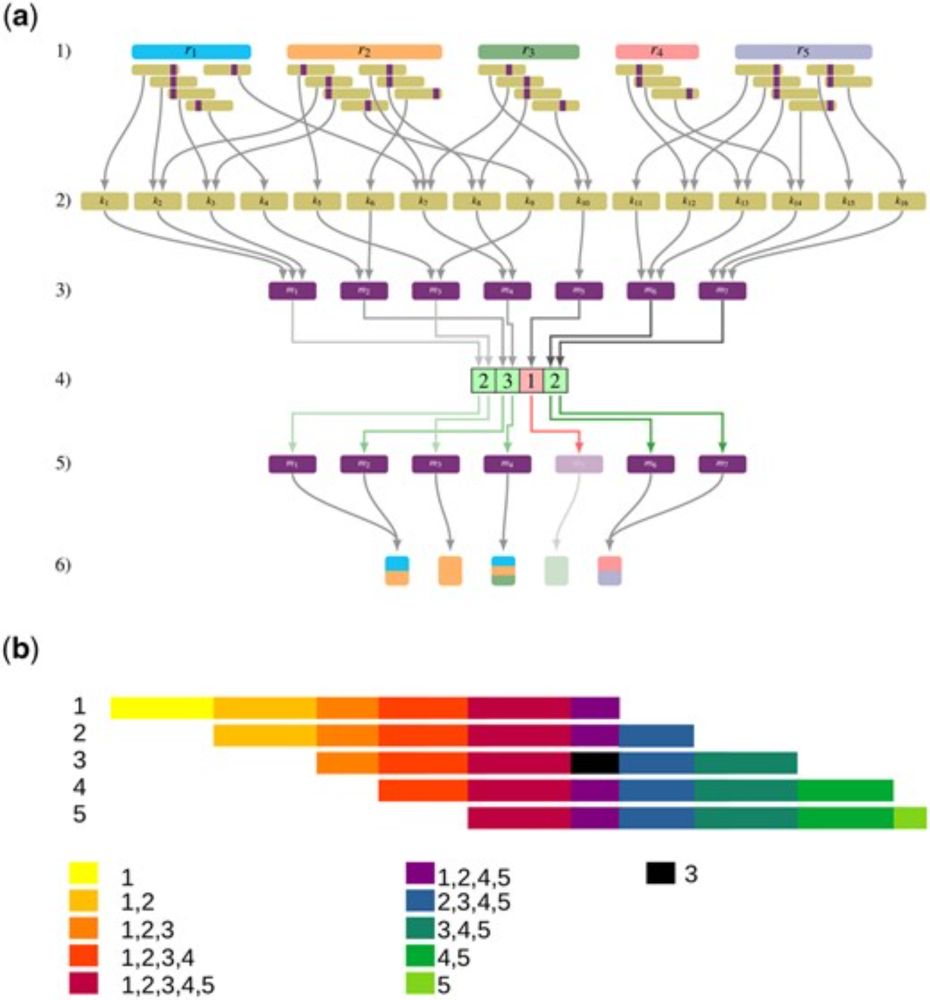
K2R: Tinted de Bruijn graphs implementation for efficient read extraction from sequencing datasets
AbstractSummary. Biological sequence analysis often relies on reference genomes, but producing accurate assemblies remains a challenge. As a result, de nov
academic.oup.com
July 5, 2025 at 9:31 AM
Paper Alert!
Our preprint on the K2R index, being able to efficiently associate kmers to the reads containing them is finally out there!
A thread!
academic.oup.com/bioinformati...
Our preprint on the K2R index, being able to efficiently associate kmers to the reads containing them is finally out there!
A thread!
academic.oup.com/bioinformati...
Reposted
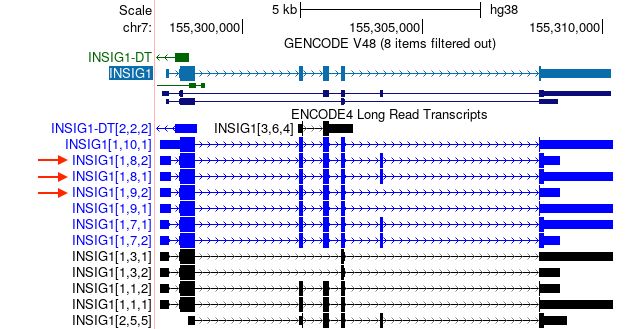
New ENCODE4 long-read RNA-seq transcripts track for hg38 and mm10. Triplets (e.g. [1,1,3]) indicate start site, exon combination, and stop site for each transcript. Enrichment scores show how these change across tissue and cell line samples.
Read more: genome.ucsc.edu/gold...
Read more: genome.ucsc.edu/gold...
July 16, 2025 at 6:27 PM
New ENCODE4 long-read RNA-seq transcripts track for hg38 and mm10. Triplets (e.g. [1,1,3]) indicate start site, exon combination, and stop site for each transcript. Enrichment scores show how these change across tissue and cell line samples.
Read more: genome.ucsc.edu/gold...
Read more: genome.ucsc.edu/gold...
Reposted
#JOBIM2025 Mathilde Girard ends the session with a simple but effective idea: re oder the reads before using an off the shelf compressor to improve compression gain

July 10, 2025 at 9:34 AM
#JOBIM2025 Mathilde Girard ends the session with a simple but effective idea: re oder the reads before using an off the shelf compressor to improve compression gain
Reposted
#JOBIM2025 @bdegardins.bsky.social presents his PhD work on Vizitig, a multi sample graph exploration tool, with a focus on RNA - this afternoon we'll do a demo on pangenomes with the same tool

July 10, 2025 at 8:44 AM
#JOBIM2025 @bdegardins.bsky.social presents his PhD work on Vizitig, a multi sample graph exploration tool, with a focus on RNA - this afternoon we'll do a demo on pangenomes with the same tool
Reposted
Paper alert!
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
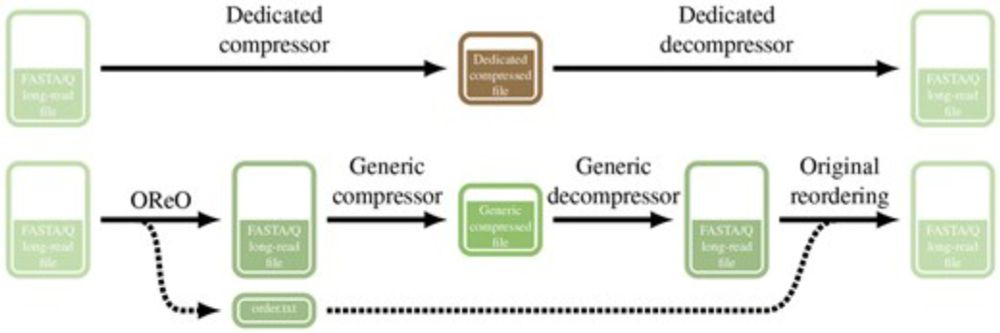
OReO: optimizing read order for practical compression
AbstractMotivation. Recent advances in high-throughput and third-generation sequencing technologies have created significant challenges in storing and mana
academic.oup.com
July 3, 2025 at 10:53 AM
Paper alert!
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
Reposted
Preprint alert from the group 🚨 super fast grep-like sequence selection
A common approach is to use k-mer indexes to identify datasets that share a significant number of k-mers with a query sequence.
This problem has been studied extensively, and I recommend the survey of @camillemrcht.bsky.social, a great introduction to the subject
genome.cshlp.org/content/31/1...
This problem has been studied extensively, and I recommend the survey of @camillemrcht.bsky.social, a great introduction to the subject
genome.cshlp.org/content/31/1...
Data structures based on k-mers for querying large collections of sequencing data sets
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
genome.cshlp.org
July 2, 2025 at 1:38 PM
Preprint alert from the group 🚨 super fast grep-like sequence selection
What a thread!
Preprint alert! 🦌
Our new abundance index, REINDEER2, is out!
It's cheap to build and update, offers tunable abundance precision at kmer level, and delivers very high query throughput.
Short thread!
www.biorxiv.org/content/10.1...
github.com/Yohan-Hernan...
Our new abundance index, REINDEER2, is out!
It's cheap to build and update, offers tunable abundance precision at kmer level, and delivers very high query throughput.
Short thread!
www.biorxiv.org/content/10.1...
github.com/Yohan-Hernan...
www.biorxiv.org
June 20, 2025 at 1:39 AM
What a thread!
Now indexing 10k RNA-seq samples with accurate counts!
REINDEER2: practical abundance index at scale https://www.biorxiv.org/content/10.1101/2025.06.16.659990v1
June 17, 2025 at 2:39 PM
Now indexing 10k RNA-seq samples with accurate counts!
Reposted
Hey yeast lovers. Do you like pangenomes?
O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻
O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻

June 11, 2025 at 2:46 PM
Hey yeast lovers. Do you like pangenomes?
O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻
O'Donnel et al. 2023 produced T2T assemblies of different strains, including phased haplotypes for yeast.
Here I selected 10 phased haplotypes and the S288C reference,
and looked for the MST28 / YAR033W gene reported to contain SVs such as indels.
👇🏻👇🏻
Reposted
A great and non technical, yet super relevant piece on implications of this scam
I've written a Substack about the #ColossalBioSci #DeExtinction #DisInformation campaign; it's focused on polluting the information landscape, rather than the underlying science (except for species concepts, I talk about them)
open.substack.com/pub/devoevom...
open.substack.com/pub/devoevom...

The Extinction of Truth
Or, a Colossal Pile of Bullshit
open.substack.com
May 14, 2025 at 3:12 PM
A great and non technical, yet super relevant piece on implications of this scam
Reposted
Just merged in an awesome new feature for xsra to support named pipes with @robp.bsky.social.
This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.
This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.

May 9, 2025 at 3:26 PM
Just merged in an awesome new feature for xsra to support named pipes with @robp.bsky.social.
This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.
This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.
Reposted
gtexr: R interface to the GTEx Portal API docs.ropensci.org/gtexr/ #rstats

Query the GTEx Portal API
A convenient R interface to the Genotype-Tissue Expression (GTEx) Portal API. The GTEx project is a comprehensive public resource for studying tissue-specific gene expression and regulation in human…
docs.ropensci.org
April 28, 2025 at 3:30 PM
gtexr: R interface to the GTEx Portal API docs.ropensci.org/gtexr/ #rstats
Reposted
J'ai rarement vu une désinformation d'une telle ampleur, de la part d'un journal soi-disant sérieux, faisant dire à une expertise de l'Anses à peu près l'inverse de ce qu'elle dit vraiment...
C'est fou.
Même si c'est désormais récurrent.
On reprend tout ? 🧵
C'est fou.
Même si c'est désormais récurrent.
On reprend tout ? 🧵
April 24, 2025 at 2:03 PM
J'ai rarement vu une désinformation d'une telle ampleur, de la part d'un journal soi-disant sérieux, faisant dire à une expertise de l'Anses à peu près l'inverse de ce qu'elle dit vraiment...
C'est fou.
Même si c'est désormais récurrent.
On reprend tout ? 🧵
C'est fou.
Même si c'est désormais récurrent.
On reprend tout ? 🧵
Reposted
This is so destructive and idiotic. mRNA vaccines are one of the most exciting opportunities to tackle diseases - from infectious disease to cancer and beyond - and these morons want to shut it down.
Exclusive: NIH officials have advised scientists to remove reference to mRNA vaccines from their grant applications, in expectation the Trump administration intends to abandon most research in the field.
By @arthurallen202.bsky.social
kffhealthnews.org/news/article...
By @arthurallen202.bsky.social
kffhealthnews.org/news/article...
Scientists Say NIH Officials Told Them To Scrub mRNA References on Grants - KFF Health News
Two senior scientists say National Institutes of Health officials advised them to remove references to mRNA vaccines in grant applications, and they fear the Trump administration will abandon a promis...
kffhealthnews.org
March 17, 2025 at 8:06 AM
This is so destructive and idiotic. mRNA vaccines are one of the most exciting opportunities to tackle diseases - from infectious disease to cancer and beyond - and these morons want to shut it down.
Reposted
correct RNAhub webserver address
rnahub.org
rnahub.org

We don't use cookies to track you - a session cookie is used exclusively for form protection (CSRF Token) only when submitting a search
rnahub.org
March 16, 2025 at 12:34 PM
correct RNAhub webserver address
rnahub.org
rnahub.org
Bioinfo PhDs from abroad and french PhDs willing to return: There is still time to apply and be part of this super innovative project on cancer transcriptomics.
Dear young investigator in cancer bioinformatics: check our postdoc position on reference-free, organ agnostic transcriptome signatures of cancer at I2BC, Gif sur Yvette.
emploi.cnrs.fr/Offres/CDD/U...
emploi.cnrs.fr/Offres/CDD/U...

March 5, 2025 at 5:21 PM
Bioinfo PhDs from abroad and french PhDs willing to return: There is still time to apply and be part of this super innovative project on cancer transcriptomics.

