Learn more at http://genome.ucsc.e...

Learn more at http://genome.ucsc.e...

The incumbent will be responsible for the Genomics Institute's complex computing infrastructure, including central and departmental systems, high-throughput storage, web systems, & cloud environments.
Apply at jobs.ucsc.edu using job code 82880.

The incumbent will be responsible for the Genomics Institute's complex computing infrastructure, including central and departmental systems, high-throughput storage, web systems, & cloud environments.
Apply at jobs.ucsc.edu using job code 82880.

genome.ucsc.edu/gold...

genome.ucsc.edu/gold...
Learn more about the release from the following news post:
genome.ucsc.edu/gold...
Learn more about the release from the following news post:
genome.ucsc.edu/gold...


Learn more about this release at:
genome.ucsc.edu/gold...

Learn more about this release at:
genome.ucsc.edu/gold...
Learn more at:
genome.ucsc.edu/gold...

Learn more at:
genome.ucsc.edu/gold...
Your feedback supports future improvements and our continued funding.

Your feedback supports future improvements and our continued funding.
Learn more at:
http://bit.ly/4lCIlLq

Learn more at:
http://bit.ly/4lCIlLq
See our news for more: bit.ly/CLSlongRead

See our news for more: bit.ly/CLSlongRead

Thanks to the Boeva Lab at ETH Zurich for creating this hub.
Thanks to the Boeva Lab at ETH Zurich for creating this hub.
Read more at:
genome.ucsc.edu/gold...
Read more at:
genome.ucsc.edu/gold...

🔗 forms.gle/s2QnMrbEYu...
🔗 forms.gle/s2QnMrbEYu...
Learn more at:
genome.ucsc.edu/gold...
Learn more at:
genome.ucsc.edu/gold...
Read more: genome.ucsc.edu/gold...
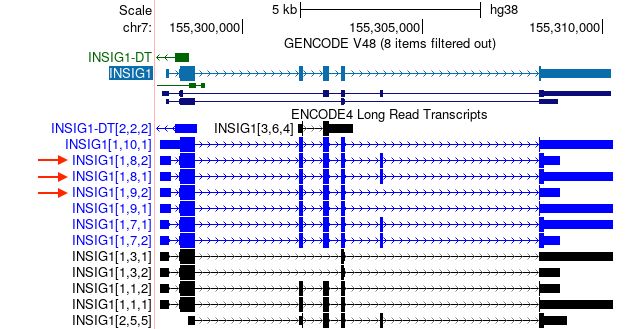
Read more: genome.ucsc.edu/gold...
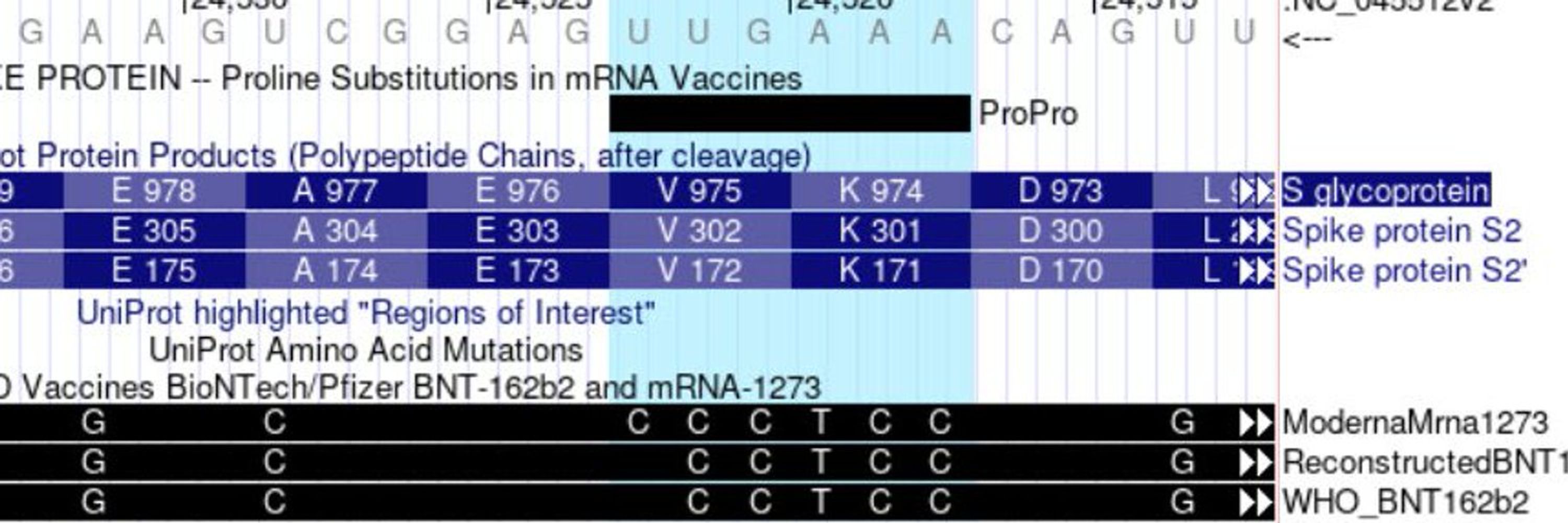
Both are aimed at interpreting the pathogenicity of variants in a clinical setting.
See our news for more: bit.ly/UCSCmutScoreMCAP

Both are aimed at interpreting the pathogenicity of variants in a clinical setting.
See our news for more: bit.ly/UCSCmutScoreMCAP
genome.ucsc.edu/gold...
genome.ucsc.edu/gold...

