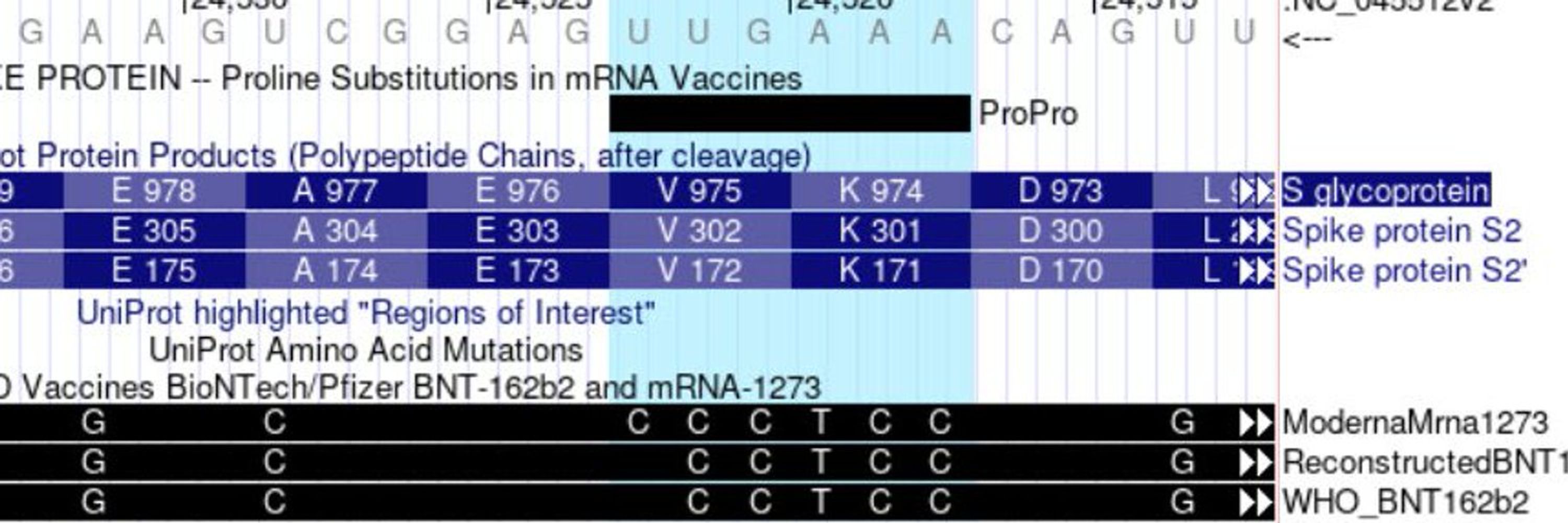


Learn more about this release at:
genome.ucsc.edu/gold...

Learn more about this release at:
genome.ucsc.edu/gold...
Learn more at:
genome.ucsc.edu/gold...

Learn more at:
genome.ucsc.edu/gold...
Learn more at:
http://bit.ly/4lCIlLq

Learn more at:
http://bit.ly/4lCIlLq
See our news for more: bit.ly/CLSlongRead

See our news for more: bit.ly/CLSlongRead


🔗 forms.gle/s2QnMrbEYu...
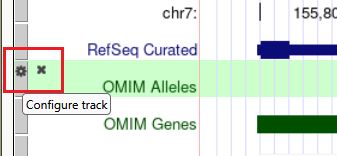
🔗 forms.gle/s2QnMrbEYu...
Read more: genome.ucsc.edu/gold...
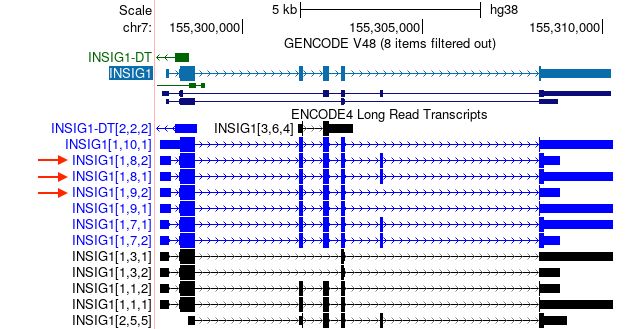
Read more: genome.ucsc.edu/gold...
Both are aimed at interpreting the pathogenicity of variants in a clinical setting.
See our news for more: bit.ly/UCSCmutScoreMCAP

Both are aimed at interpreting the pathogenicity of variants in a clinical setting.
See our news for more: bit.ly/UCSCmutScoreMCAP
July 7th marks the 25th anniversary of the human genome going online and the start of the UCSC Genome Browser.
Then vs. now, we have 165k monthly visitors, and our codebase is over three million lines of code.
See our news for more: bit.ly/genomeBrowser...
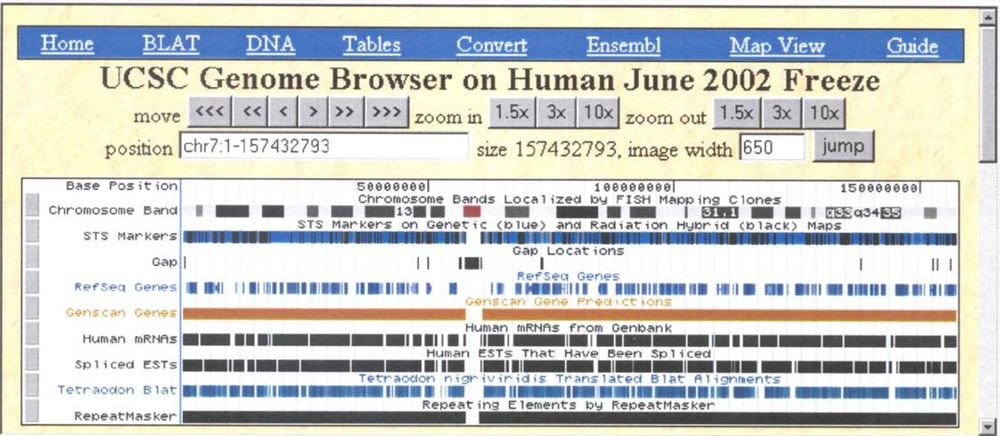
July 7th marks the 25th anniversary of the human genome going online and the start of the UCSC Genome Browser.
Then vs. now, we have 165k monthly visitors, and our codebase is over three million lines of code.
See our news for more: bit.ly/genomeBrowser...
See our news for more: bit.ly/genomeBrowser...

See our news for more: bit.ly/genomeBrowser...
Learn more at http://genome.ucsc.e....

Learn more at http://genome.ucsc.e....
Learn more at:
genome.ucsc.edu/gold...

Learn more at:
genome.ucsc.edu/gold...
See our news for more: bit.ly/ExonRelevance...

See our news for more: bit.ly/ExonRelevance...
The pext track shows @gnomad-project.bsky.social data in combination with GTExV10 to compute a metric quantifying isoform expression for variants.

The pext track shows @gnomad-project.bsky.social data in combination with GTExV10 to compute a metric quantifying isoform expression for variants.
Learn more at genome.ucsc.edu/gold....

Learn more at genome.ucsc.edu/gold....
See our news for more: genome.ucsc.edu/gold...

See our news for more: genome.ucsc.edu/gold...
Learn more at genome.ucsc.edu/gold....

Learn more at genome.ucsc.edu/gold....

Learn more about this release at:
genome.ucsc.edu/gold...

Learn more about this release at:
genome.ucsc.edu/gold...

Learn more at:
genome.ucsc.edu/gold...

Learn more at:
genome.ucsc.edu/gold...



