
Elena Rivas
@rivaselenarivas.bsky.social
computational biologist. algorithms for genomics. worried about sustainability: from personal 2 country 2 planet. 🧪🧬| http://rivaslab.org
Pinned

All-at-once RNA folding with 3D motif prediction framed by evolutionary information - Nature Methods
Structural RNAs exhibit a vast array of recurrent short three-dimensional (3D) elements found in loop regions involving non-Watson–Crick interactions that help arrange canonical double helices into tertiary structures. Here we present CaCoFold-R3D, a probabilistic grammar that predicts these RNA 3D motifs (also termed modules) jointly with RNA secondary structure over a sequence or alignment. CaCoFold-R3D uses evolutionary information present in an RNA alignment to reliably identify canonical helices (including pseudoknots) by covariation. Here we further introduce the R3D grammars, which also exploit helix covariation that constrains the positioning of the mostly noncovarying RNA 3D motifs. Our method runs predictions over an almost-exhaustive list of over 50 known RNA motifs (‘everything’). Motifs can appear in any nonhelical loop region (including three-way, four-way and higher junctions) (‘everywhere’). All structural motifs as well as the canonical helices are arranged into one single structure predicted by one single joint probabilistic grammar (‘all-at-once’). Our results demonstrate that CaCoFold-R3D is a valid alternative for predicting the all-residue interactions present in a RNA 3D structure. CaCoFold-R3D is fast and easily customizable for novel motif discovery and shows promising value both as a strong input for deep learning approaches to all-atom structure prediction as well as toward guiding RNA design as drug targets for therapeutic small molecules.
link.springer.com
Integrated prediction of RNA secondary structure jointly with 3D motifs and pseudoknots guided by evolutionary information.
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
RNA BENASQUE
July 05-17 2026
July 05-17 2026
November 1, 2025 at 2:02 AM
RNA BENASQUE
July 05-17 2026
July 05-17 2026
Registration to the next RNA BENASQUE meeting July 05-17 2026 is open.
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
Computational Approaches to RNA Structure and Function
benasque.org
November 1, 2025 at 1:03 AM
Registration to the next RNA BENASQUE meeting July 05-17 2026 is open.
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
Reposted by Elena Rivas
Please join us for our next seminar, featuring Danny IncaRNAto, PhD: “Discovery of RNA regulatory structural switches via ensemble mapping”
@incarnatolab.bsky.social #rna #casprnasig #rnastructure
@incarnatolab.bsky.social #rna #casprnasig #rnastructure

October 10, 2025 at 5:34 PM
Please join us for our next seminar, featuring Danny IncaRNAto, PhD: “Discovery of RNA regulatory structural switches via ensemble mapping”
@incarnatolab.bsky.social #rna #casprnasig #rnastructure
@incarnatolab.bsky.social #rna #casprnasig #rnastructure
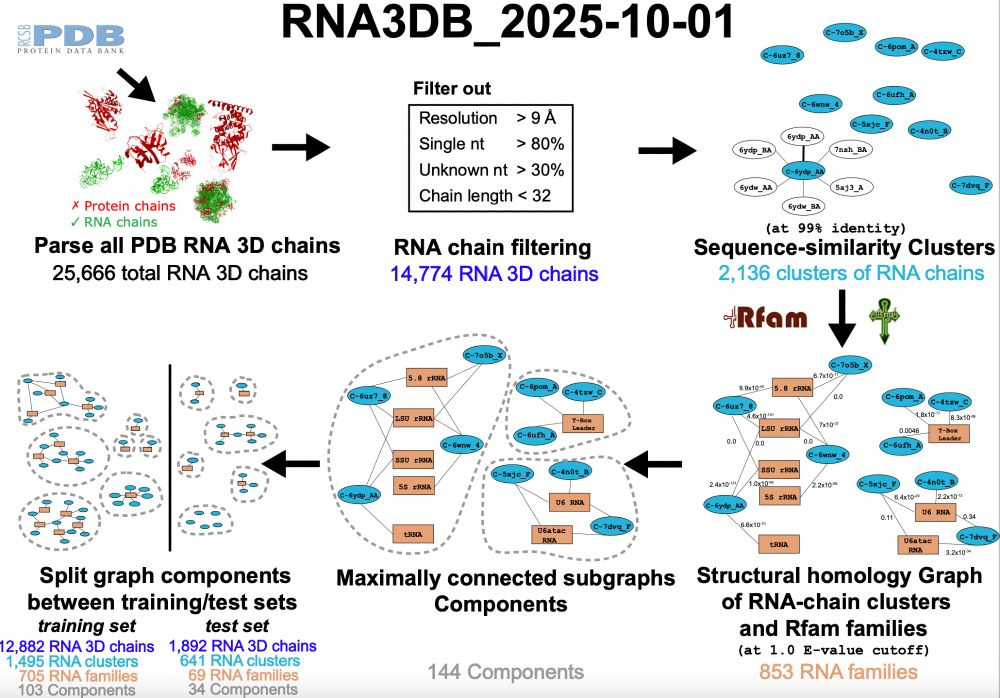
RNA3DB 2025-10-01 release
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
October 10, 2025 at 3:51 PM
RNA3DB 2025-10-01 release
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
Reposted by Elena Rivas
RNA Structure beyond Canonical Base Pairs Guided by Evolution 🧪🧬 #AcademicSky #higherEd
www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social
www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social

RNA Structure beyond Canonical Base Pairs Guided by Evolution - Harvard University - Department of Molecular & Cellular Biology
In addition to messenger RNA (mRNA) that provide the code for proteins, there are other RNAs that exert their function just as RNA, usually referred to as non-coding […]
www.mcb.harvard.edu
October 7, 2025 at 10:37 PM
RNA Structure beyond Canonical Base Pairs Guided by Evolution 🧪🧬 #AcademicSky #higherEd
www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social
www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social
CaCoFold-R3D: a probabilistic model that predicts RNA 3D motifs and secondary structure using evolutionary information.
www.nature.com/articles/s41...
www.nature.com/articles/s41...

All-at-once RNA folding with 3D motif prediction framed by evolutionary information - Nature Methods
CaCoFold-R3D is a probabilistic model that simultaneously predicts the RNA 3D motifs jointly with the secondary structure in a structural RNA using evolutionary information.
www.nature.com
October 3, 2025 at 2:18 PM
Reposted by Elena Rivas
Laser ablation of imaged surface layers under adaptive control enables volumetric imaging of heterogeneous tissue like skull and brain.
www.nature.com/articles/s41...
www.nature.com/articles/s41...

October 3, 2025 at 1:31 PM
Laser ablation of imaged surface layers under adaptive control enables volumetric imaging of heterogeneous tissue like skull and brain.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Integrated prediction of RNA secondary structure jointly with 3D motifs and pseudoknots guided by evolutionary information.
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...

All-at-once RNA folding with 3D motif prediction framed by evolutionary information - Nature Methods
Structural RNAs exhibit a vast array of recurrent short three-dimensional (3D) elements found in loop regions involving non-Watson–Crick interactions that help arrange canonical double helices into tertiary structures. Here we present CaCoFold-R3D, a probabilistic grammar that predicts these RNA 3D motifs (also termed modules) jointly with RNA secondary structure over a sequence or alignment. CaCoFold-R3D uses evolutionary information present in an RNA alignment to reliably identify canonical helices (including pseudoknots) by covariation. Here we further introduce the R3D grammars, which also exploit helix covariation that constrains the positioning of the mostly noncovarying RNA 3D motifs. Our method runs predictions over an almost-exhaustive list of over 50 known RNA motifs (‘everything’). Motifs can appear in any nonhelical loop region (including three-way, four-way and higher junctions) (‘everywhere’). All structural motifs as well as the canonical helices are arranged into one single structure predicted by one single joint probabilistic grammar (‘all-at-once’). Our results demonstrate that CaCoFold-R3D is a valid alternative for predicting the all-residue interactions present in a RNA 3D structure. CaCoFold-R3D is fast and easily customizable for novel motif discovery and shows promising value both as a strong input for deep learning approaches to all-atom structure prediction as well as toward guiding RNA design as drug targets for therapeutic small molecules.
link.springer.com
October 3, 2025 at 12:42 PM
Integrated prediction of RNA secondary structure jointly with 3D motifs and pseudoknots guided by evolutionary information.
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
Reposted by Elena Rivas
Vaccines have one of the highest reward to risk ratios of any medical intervention in human history. This sociopathic conspiracy theorist is literally killing people, causing the deaths by vaccine-preventable diseases of people who could have lived.
there is no question in my mind that rfk jr is the most dangerous person in this administration and that his eugenicist ideology threatens the lives of millions of people www.advocate.com/politics/dem...

RFK Jr.’s damage to the CDC is ‘past the point of no return,’ Dr. Demetre Daskalakis warns
“The CDC you knew is over,” the infectious diseases doctor told The Advocate. “Unless someone takes radical action, there is nothing there that can be salvaged.”
www.advocate.com
August 30, 2025 at 12:24 AM
Vaccines have one of the highest reward to risk ratios of any medical intervention in human history. This sociopathic conspiracy theorist is literally killing people, causing the deaths by vaccine-preventable diseases of people who could have lived.
Reposted by Elena Rivas
New York Times story with profiles of researchers whose grants were terminated.
[Gift Link]
www.nytimes.com/2025/08/24/o...
[Gift Link]
www.nytimes.com/2025/08/24/o...

Opinion | America First? Not When It Comes to Your Health.
www.nytimes.com
August 24, 2025 at 1:08 PM
New York Times story with profiles of researchers whose grants were terminated.
[Gift Link]
www.nytimes.com/2025/08/24/o...
[Gift Link]
www.nytimes.com/2025/08/24/o...
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
doi.org
August 2, 2025 at 8:39 PM
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
Reposted by Elena Rivas
The neighborhood is getting ready for tomorrow

June 13, 2025 at 4:56 PM
The neighborhood is getting ready for tomorrow
Reposted by Elena Rivas
Kseniia Petrova walked out of the Boston federal courthouse this afternoon on pre-trial release. My writeup for @cambridgeday.com
www.cambridgeday.com/2025/06/12/p...
www.cambridgeday.com/2025/06/12/p...

Petrova released on bail from federal detention, walks out of courthouse; indictment still possible - Cambridge Day
Harvard researcher Kseniia Petrova was released on bail after a 10-minute hearing in her criminal smuggling case – of frog embryos meant for scientific use.
www.cambridgeday.com
June 12, 2025 at 7:48 PM
Kseniia Petrova walked out of the Boston federal courthouse this afternoon on pre-trial release. My writeup for @cambridgeday.com
www.cambridgeday.com/2025/06/12/p...
www.cambridgeday.com/2025/06/12/p...
Reposted by Elena Rivas
Reminder: Nobel-prize winning PCR (1983), used in basically all genetic tech today, was only possible because of extremophile bacterium discovered in 1964 in Yellowstone funded by a small ~$80k NSF grant with no obvious application at the time. #science 🧪
www.richmondscientific.com/how-a-discov...
www.richmondscientific.com/how-a-discov...

How a discovery in Yellowstone National Park led to the development of PCR - Richmond Scientific
A discovery in Yellowstone National Park led to the development of PCR, the gold-standard COVID-19 tests used to fight the global pandemic.
www.richmondscientific.com
June 8, 2025 at 9:09 PM
Reminder: Nobel-prize winning PCR (1983), used in basically all genetic tech today, was only possible because of extremophile bacterium discovered in 1964 in Yellowstone funded by a small ~$80k NSF grant with no obvious application at the time. #science 🧪
www.richmondscientific.com/how-a-discov...
www.richmondscientific.com/how-a-discov...
Important validation of our covariation-based screens for conserved structural RNAs. academic.oup.com/nar/article/...
work lead (both computational and experimental) by then undergraduate, William Gao
work lead (both computational and experimental) by then undergraduate, William Gao
June 9, 2025 at 1:30 AM
Important validation of our covariation-based screens for conserved structural RNAs. academic.oup.com/nar/article/...
work lead (both computational and experimental) by then undergraduate, William Gao
work lead (both computational and experimental) by then undergraduate, William Gao
Reposted by Elena Rivas
Another fantastic explanation of how basic science helps people and society.
Northwestern scientist Julius Lucks makes an eloquent appeal for why research funding matters to us all. tinyurl.com/yjbk5zca #syntheticbiology #research #northwestern #funding

What’s next for Northwestern researchers amid funding cuts and work stoppages?
Northwestern researchers received notices from the Department of Defense to stop their work immediately.
tinyurl.com
June 8, 2025 at 1:19 AM
Another fantastic explanation of how basic science helps people and society.
Reposted by Elena Rivas
This NYT analysis is incomplete, because the underlying data provided by HHS is inaccurate. It does not include the mass termination of all federal funding to Harvard.
www.nytimes.com/interactive/...
www.nytimes.com/interactive/...

Here Are the Nearly 2,500 Medical Research Grants Canceled or Delayed by Trump
Some cuts have been starkly visible, but the country’s medical grant-making machinery has also radically transformed outside the public eye.
www.nytimes.com
June 4, 2025 at 3:09 PM
This NYT analysis is incomplete, because the underlying data provided by HHS is inaccurate. It does not include the mass termination of all federal funding to Harvard.
www.nytimes.com/interactive/...
www.nytimes.com/interactive/...
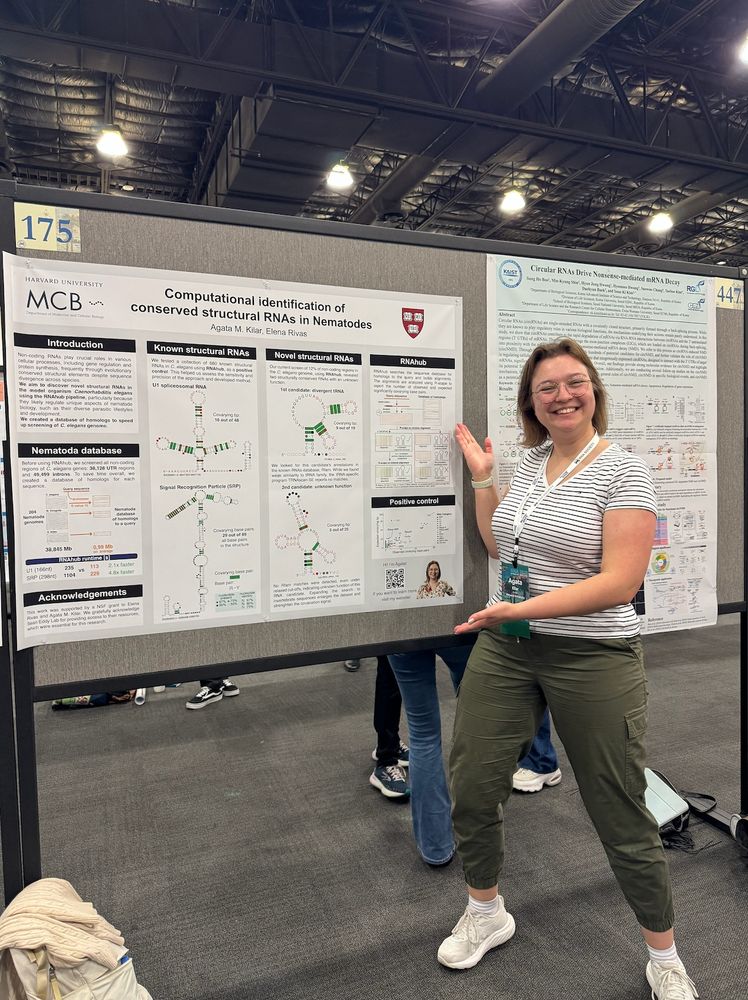
The lab is at the RNA Society meeting #RNA25 #RNA2025
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
May 29, 2025 at 4:21 PM
The lab is at the RNA Society meeting #RNA25 #RNA2025
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
Reposted by Elena Rivas
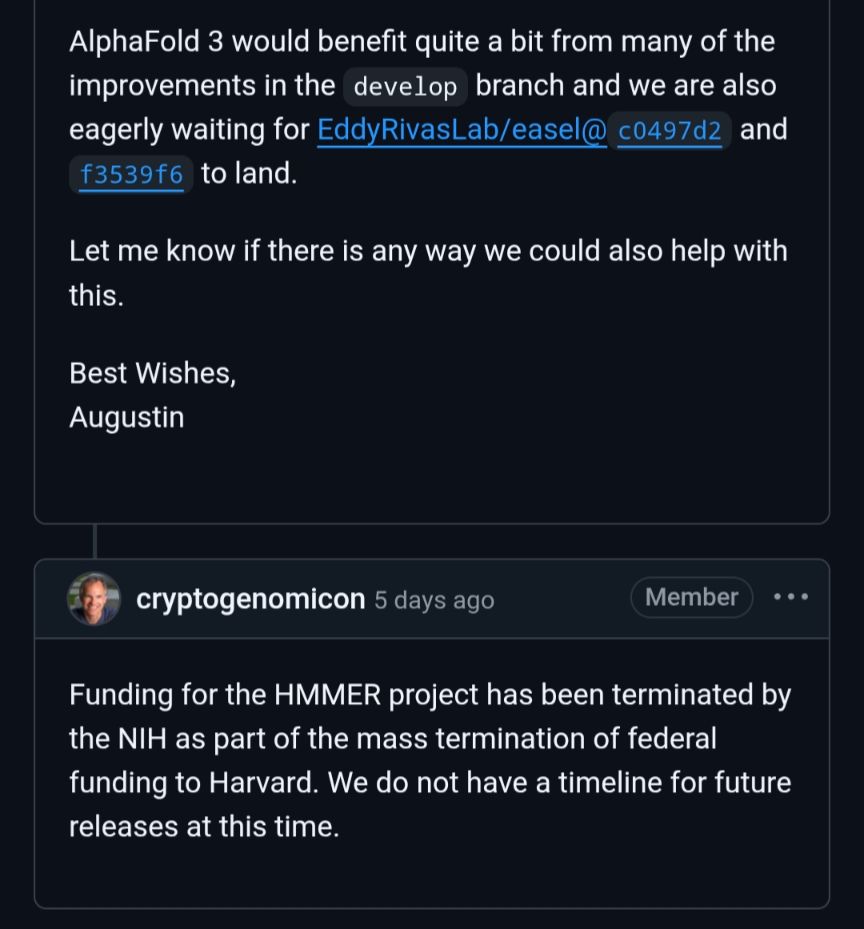
The war on science in the US is already having an effect on private sector research like AlphaFold. Bears repeating but the private sector builds on top of things created by academic research for the public good. This hurts everyone.
May 28, 2025 at 10:18 AM
The war on science in the US is already having an effect on private sector research like AlphaFold. Bears repeating but the private sector builds on top of things created by academic research for the public good. This hurts everyone.
Reposted by Elena Rivas
The $700M/year of federal funding to Harvard (not $3B or other fictional numbers) is for research labs, not for the college. Trade schools are great, but taking science funding and giving it to trade schools shows a misunderstanding of how the US has funded scientific research since WWII.
May 26, 2025 at 1:33 PM
The $700M/year of federal funding to Harvard (not $3B or other fictional numbers) is for research labs, not for the college. Trade schools are great, but taking science funding and giving it to trade schools shows a misunderstanding of how the US has funded scientific research since WWII.
Reposted by Elena Rivas
(1/7) Very excited to share my first PhD preprint on the interactions of two of my favorite mobile genetic elements: phages and group II introns!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Prevalence of Group II Introns in Phage Genomes
Although bacteriophage genomes are under strong selective pressure for high coding density, they are still frequently invaded by mobile genetic elements (MGEs). Group II introns are MGEs that reduce h...
www.biorxiv.org
May 24, 2025 at 7:20 PM
(1/7) Very excited to share my first PhD preprint on the interactions of two of my favorite mobile genetic elements: phages and group II introns!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Elena Rivas
On a day when all research grants at Harvard were terminated, and in a context where Congress seems to broadly accept the notion that destroying science and health research in the US is fine, it’s worth noting that this is not what the public wants.
The American public is broadly supportive of maintaining federal science and medical research funding, and against punishing universities by taking their funding away, according to a new poll:

Few support punitive funding cuts to colleges and universities - AP-NORC
Most of the public believes that colleges and universities make significant contributions to scientific and technological advancement and think their federal research funding should be maintained.
apnorc.org
May 15, 2025 at 9:54 PM
On a day when all research grants at Harvard were terminated, and in a context where Congress seems to broadly accept the notion that destroying science and health research in the US is fine, it’s worth noting that this is not what the public wants.
All funding that supports my lab including myself terminated, just now.

May 15, 2025 at 9:09 PM
All funding that supports my lab including myself terminated, just now.
Reposted by Elena Rivas








