Marcel Schulz
@themarcelschulz.bsky.social
Director of Institute for Computational Genomic Medicine at Goethe University Frankfurt https://cgm.uni-frankfurt.de/
Reposted by Marcel Schulz
In a new work with Joseph Rich and Conrad Oakes we tackle the problem of how to best organize alluvial plots. We formalize two optimization problems and develop a solution for them based on the neighbornet algorithm, implemented in the program wompwomp: github.com/pachterlab/w...

September 5, 2025 at 12:20 PM
In a new work with Joseph Rich and Conrad Oakes we tackle the problem of how to best organize alluvial plots. We formalize two optimization problems and develop a solution for them based on the neighbornet algorithm, implemented in the program wompwomp: github.com/pachterlab/w...
Reposted by Marcel Schulz
🚀 Two exciting days of Computational Biology at the Cardio-Pulmonary Institute in Bad Nauheim @cpi-exstra.bsky.social ! 🔬 Workshop on single-cell analysis + the CPI repository data ressource; 💡 Hackathon together with the Schulz Lab @themarcelschulz.bsky.social
Collaboration, coding & discovery
Collaboration, coding & discovery

August 28, 2025 at 12:07 PM
🚀 Two exciting days of Computational Biology at the Cardio-Pulmonary Institute in Bad Nauheim @cpi-exstra.bsky.social ! 🔬 Workshop on single-cell analysis + the CPI repository data ressource; 💡 Hackathon together with the Schulz Lab @themarcelschulz.bsky.social
Collaboration, coding & discovery
Collaboration, coding & discovery
Reposted by Marcel Schulz
🎉 To mark the fourth extension of the CPI, we celebrated our success at the @mpi-hlr.bsky.social 🥳 We are delighted about the renewed funding and would like to thank everyone who made this step possible! Read more in the latest issue of "Synapse": www.unimedizin-ffm.de/fileadmin/re...

August 4, 2025 at 6:56 AM
🎉 To mark the fourth extension of the CPI, we celebrated our success at the @mpi-hlr.bsky.social 🥳 We are delighted about the renewed funding and would like to thank everyone who made this step possible! Read more in the latest issue of "Synapse": www.unimedizin-ffm.de/fileadmin/re...
Reposted by Marcel Schulz
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
doi.org
August 2, 2025 at 8:39 PM
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
Reposted by Marcel Schulz
A resource of RNA-binding protein motifs across eukaryotes reveals evolutionary dynamics and gene-regulatory function go.nature.com/4mc1SmQ
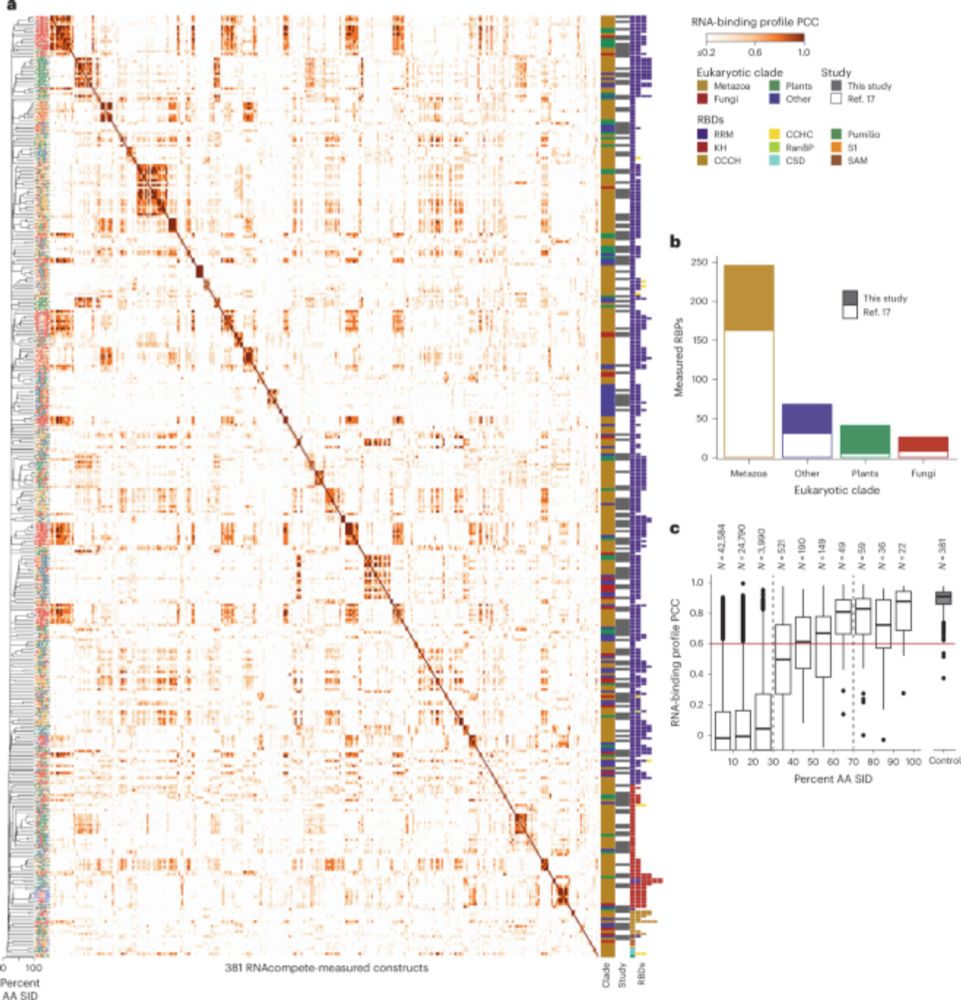
A resource of RNA-binding protein motifs across eukaryotes reveals evolutionary dynamics and gene-regulatory function - Nature Biotechnology
RNA-binding motifs in eukaryotic proteins are presented in a comprehensive resource.
go.nature.com
July 25, 2025 at 3:46 PM
A resource of RNA-binding protein motifs across eukaryotes reveals evolutionary dynamics and gene-regulatory function go.nature.com/4mc1SmQ
Reposted by Marcel Schulz
Multi-omics with A.I. analytics demonstrates critical role of the gut microbiome and immune response and enables accurate diagnosis of ME/CFS, may also be applicable to Long Covid.
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
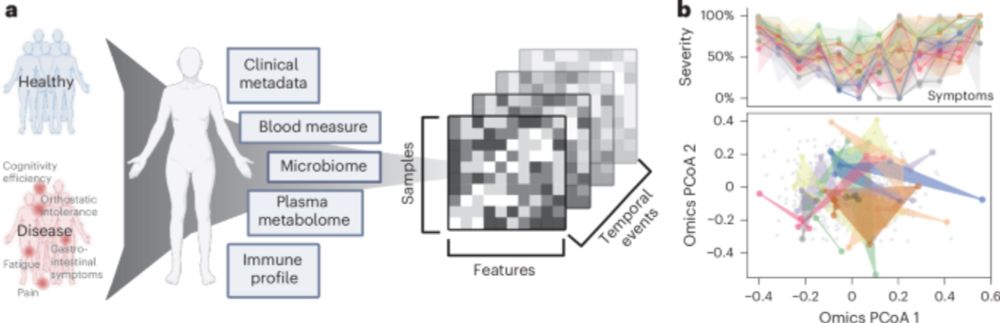
AI-driven multi-omics modeling of myalgic encephalomyelitis/chronic fatigue syndrome - Nature Medicine
Analysis of a deeply phenotyped, longitudinal cohort of 249 participants elucidates the network of concurrent molecular and macroscopic patterns related to insurgence and course of ME/CFS.
www.nature.com
July 25, 2025 at 4:08 PM
Multi-omics with A.I. analytics demonstrates critical role of the gut microbiome and immune response and enables accurate diagnosis of ME/CFS, may also be applicable to Long Covid.
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
Reposted by Marcel Schulz
Our collaboration with Tony Mustoe's lab us out today. MPRA libraries and possible cryptic splicing in MPRA reporters, or reporters in general. The additional novelty here is that such events are controlled or influenced by AU-rich sequences.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
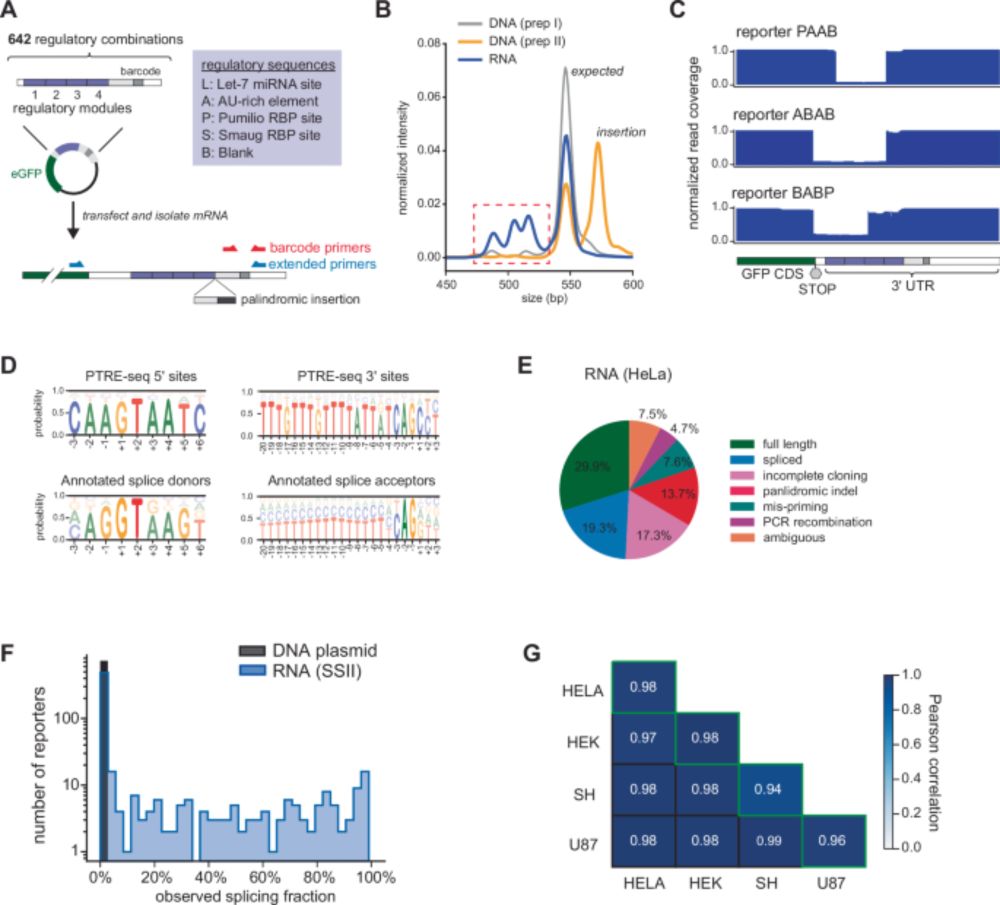
U-rich elements drive pervasive cryptic splicing in 3’ UTR massively parallel reporter assays - Nature Communications
Massively parallel reporter assays (MPRAs) are powerful technologies for measuring the impact of non-coding sequences on gene expression. Here, the authors demonstrate that MPRA reporters often underg...
www.nature.com
July 25, 2025 at 10:17 AM
Our collaboration with Tony Mustoe's lab us out today. MPRA libraries and possible cryptic splicing in MPRA reporters, or reporters in general. The additional novelty here is that such events are controlled or influenced by AU-rich sequences.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Marcel Schulz
Very excited that our most significant work, a collaboration w/ Dr. Can Cenik at UT Austin on translational gene regulation, was finally published in Nature Biotechnology in a dual set of studies:
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
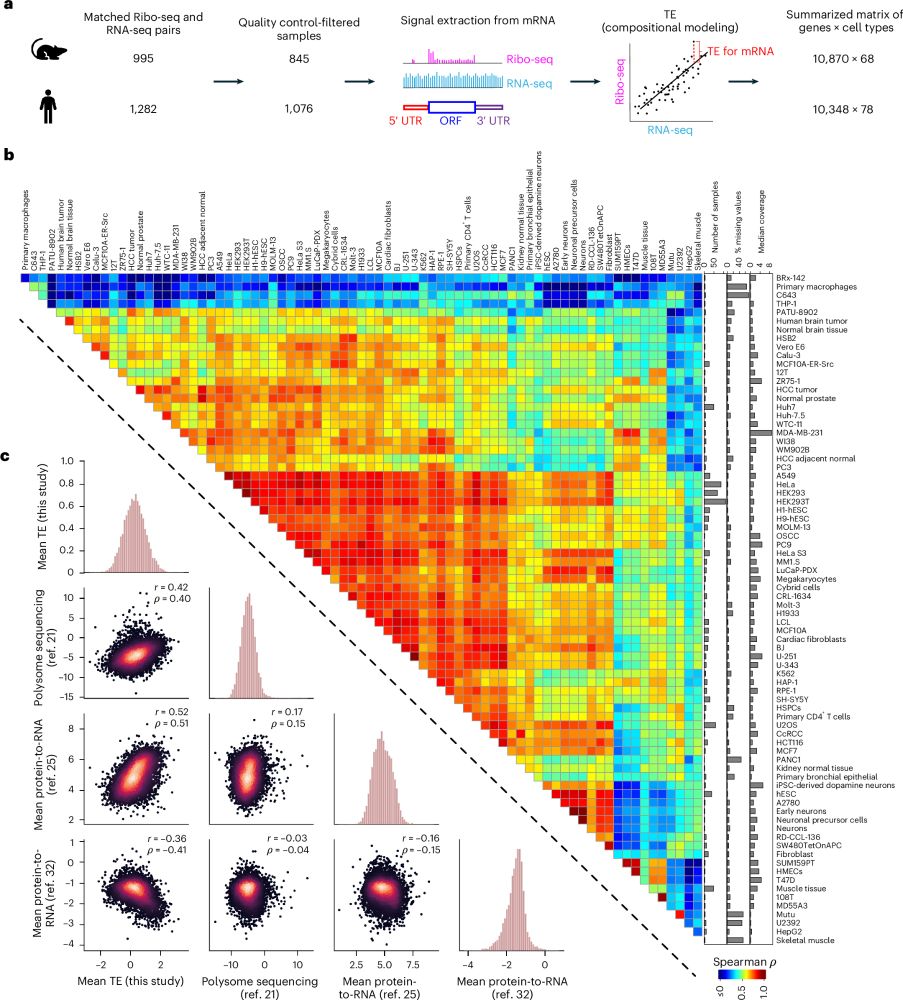
Predicting the translation efficiency of messenger RNA in mammalian cells
Nature Biotechnology - A deep convolutional neural network model predicts the influence of the full-length mRNA sequence on translation efficiency.
url.de.m.mimecastprotect.com
July 25, 2025 at 1:37 PM
Very excited that our most significant work, a collaboration w/ Dr. Can Cenik at UT Austin on translational gene regulation, was finally published in Nature Biotechnology in a dual set of studies:
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Reposted by Marcel Schulz
And BIG thanks to #RegSys track organizers @abardet.bsky.social A. Emad J. Welch R. Gordan @xiuweizhang.bsky.social R. Beekman @martamele.bsky.social @anniquec.bsky.social @shaunmahony.bsky.social @ferhatay.bsky.social and co-chairs @alemedinarivera.bsky.social and @themarcelschulz.bsky.social
July 24, 2025 at 7:00 PM
And BIG thanks to #RegSys track organizers @abardet.bsky.social A. Emad J. Welch R. Gordan @xiuweizhang.bsky.social R. Beekman @martamele.bsky.social @anniquec.bsky.social @shaunmahony.bsky.social @ferhatay.bsky.social and co-chairs @alemedinarivera.bsky.social and @themarcelschulz.bsky.social
Reposted by Marcel Schulz
What a fantastic #ISMBECCB2025 conference it has been, with an outstanding #RegSys @iscb-regsys.bsky.social COSI track!
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
July 24, 2025 at 7:00 PM
What a fantastic #ISMBECCB2025 conference it has been, with an outstanding #RegSys @iscb-regsys.bsky.social COSI track!
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
Reposted by Marcel Schulz
🧬 New @emblebi.bsky.social service beta launched:
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org

July 23, 2025 at 10:06 AM
🧬 New @emblebi.bsky.social service beta launched:
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org
Last talk at #regsys today is a keynote by Mafalda Dias about modelling sequence variation in Evolution with insights into diseases #ismbeccb2025

July 23, 2025 at 4:23 PM
Last talk at #regsys today is a keynote by Mafalda Dias about modelling sequence variation in Evolution with insights into diseases #ismbeccb2025
Reposted by Marcel Schulz
First talk of the last session of RegSys day 1 at #ismb2025 is from Jacob Schreiber on how we read the book of DNA
Spoiler: regulation is key
Spoiler: regulation is key

July 23, 2025 at 3:43 PM
First talk of the last session of RegSys day 1 at #ismb2025 is from Jacob Schreiber on how we read the book of DNA
Spoiler: regulation is key
Spoiler: regulation is key
Laura Gunsalus talks about design of small native regulatory elements using #ML #ismbeccb2015 #regsys

July 23, 2025 at 4:03 PM
Laura Gunsalus talks about design of small native regulatory elements using #ML #ismbeccb2015 #regsys
Last talk in this #regsys session is Jishnu Das who talks about regulatory programs that determine B-cell fates #ismbeccb2025

July 23, 2025 at 2:43 PM
Last talk in this #regsys session is Jishnu Das who talks about regulatory programs that determine B-cell fates #ismbeccb2025
Tatevik Jalatyan will talk about a new pipeline for estimating cell cycle phase from single cell data #ismbeccb2025 #regsys

July 23, 2025 at 2:24 PM
Tatevik Jalatyan will talk about a new pipeline for estimating cell cycle phase from single cell data #ismbeccb2025 #regsys
Surag Nair talks about a general framework to combine different paradigms of DL genome models #ismbeccb2025 #regsys

July 23, 2025 at 2:03 PM
Surag Nair talks about a general framework to combine different paradigms of DL genome models #ismbeccb2025 #regsys
Vitalii Kleshchenikow about a biophysical model for learning gene regulation from single cell data #ismbeccb2025 #regsys @oliverstegle.bsky.social

July 23, 2025 at 1:45 PM
Vitalii Kleshchenikow about a biophysical model for learning gene regulation from single cell data #ismbeccb2025 #regsys @oliverstegle.bsky.social
#regsys session is overflowing at #ismbeccb2025 consider to go to rooms 6 and 7 for video broadcast if full
July 23, 2025 at 1:40 PM
#regsys session is overflowing at #ismbeccb2025 consider to go to rooms 6 and 7 for video broadcast if full
Laura Rumpf talks about the MetaFR pipeline to learn gene regulation models from unpaired and paired scATAC and scRNA data #ismbeccb2025 #regsys #schulzlab @dzhk.de @sfb1531.bsky.social

July 23, 2025 at 1:21 PM
Laura Rumpf talks about the MetaFR pipeline to learn gene regulation models from unpaired and paired scATAC and scRNA data #ismbeccb2025 #regsys #schulzlab @dzhk.de @sfb1531.bsky.social
Refreshed after lunch we start with Oluwatosin Oluwadare to introduce a new approach for single cell HiC analysis #ismbeccb2025 #regsys
July 23, 2025 at 1:04 PM
Refreshed after lunch we start with Oluwatosin Oluwadare to introduce a new approach for single cell HiC analysis #ismbeccb2025 #regsys
Last speaker before lunch is Christophe Vroland to talk about confounders in Rna-Dna interaction data analysis #regsys #ismbeccb2025

July 23, 2025 at 12:01 PM
Last speaker before lunch is Christophe Vroland to talk about confounders in Rna-Dna interaction data analysis #regsys #ismbeccb2025
Laura Hinojosa talks about TFs that regulate replication timing
#RegSys #ismbeccb2025 @ferhatay.bsky.social
#RegSys #ismbeccb2025 @ferhatay.bsky.social

July 23, 2025 at 11:56 AM
Laura Hinojosa talks about TFs that regulate replication timing
#RegSys #ismbeccb2025 @ferhatay.bsky.social
#RegSys #ismbeccb2025 @ferhatay.bsky.social
Next is
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social

July 23, 2025 at 11:50 AM
Next is
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social
First flash talk session starts with @aryankamal.bsky.social who represents his analysis about understanding transcriptional regulation in blood generation #regsys #ismbeccb2025

July 23, 2025 at 11:45 AM
First flash talk session starts with @aryankamal.bsky.social who represents his analysis about understanding transcriptional regulation in blood generation #regsys #ismbeccb2025

