
Latest from the lab. The conclusion is in the title!
Basically, we found that the KMN complex (outer kinetochore) is fully conserved between plants and fungi/animals, showing deep origin. (reminder, you are closer to a mushroom than a mushroom is to a plant.)

Latest from the lab. The conclusion is in the title!
Basically, we found that the KMN complex (outer kinetochore) is fully conserved between plants and fungi/animals, showing deep origin. (reminder, you are closer to a mushroom than a mushroom is to a plant.)
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

rdcu.be/eLVts

rdcu.be/eLVts
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
www.nature.com/articles/s41...
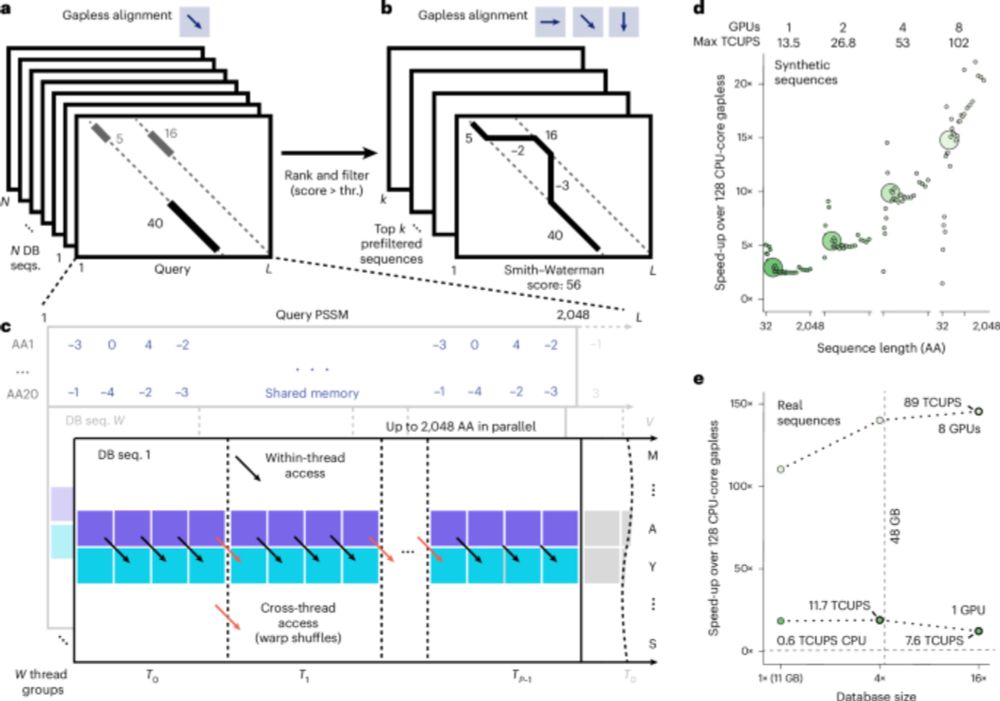
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
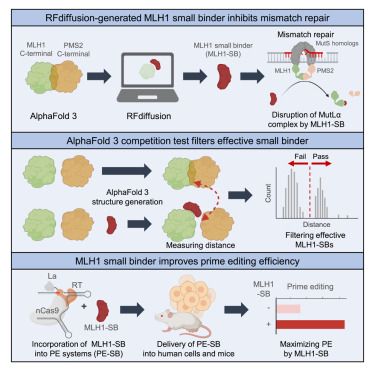
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
Chai-2, a next-gen multimodal generative model, is shaking up protein design. With a 16% hit rate in fully de novo antibody design—over 100 times better than previous methods—it’s making drug discovery faster and more precise than ever.
Chai-2, a next-gen multimodal generative model, is shaking up protein design. With a 16% hit rate in fully de novo antibody design—over 100 times better than previous methods—it’s making drug discovery faster and more precise than ever.
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
@yiweichang.bsky.social www.yiweichanglab.org @jiwasa.bsky.social #virology #Influenza #Cryo-EM #StructuralBiology #RNA #polymerase
@yiweichang.bsky.social www.yiweichanglab.org @jiwasa.bsky.social #virology #Influenza #Cryo-EM #StructuralBiology #RNA #polymerase
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.sciencedirect.com/special-issu...
www.sciencedirect.com/special-issu...
🧬🧶👩🔬👩💻 Collaborative and interdisciplinary work, together with the groups of Ankur Dalia and J. Pablo Radicella
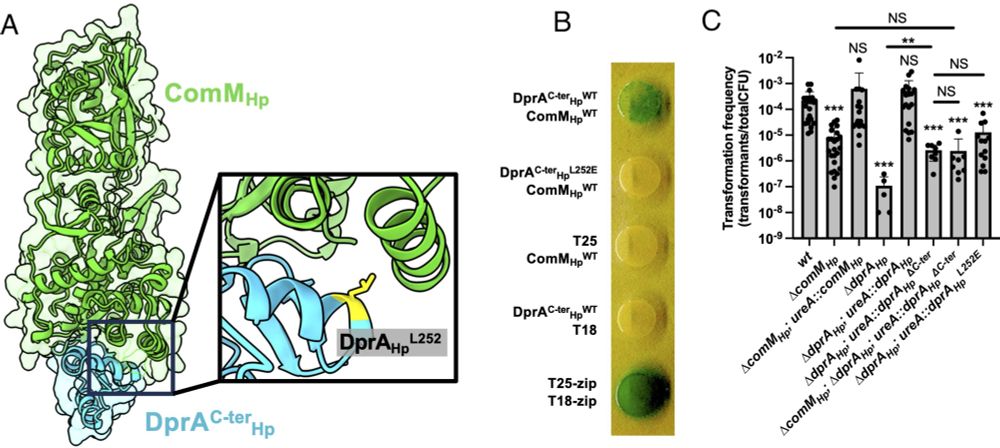
🧬🧶👩🔬👩💻 Collaborative and interdisciplinary work, together with the groups of Ankur Dalia and J. Pablo Radicella
meiosis.cornell.edu/mayosis2025/...
The #Mayosis25 seminar series will open with a tribute to Scott Hawley by @jeffsekelsky.bsky.social.
Congrats to all the selected speakers and we look forward to hearing about your fantastic science !
#Meiosis4ever
meiosis.cornell.edu/mayosis2025/...
The #Mayosis25 seminar series will open with a tribute to Scott Hawley by @jeffsekelsky.bsky.social.
Congrats to all the selected speakers and we look forward to hearing about your fantastic science !
#Meiosis4ever


