
Chosen by JAMA editors, the inaugural roundup highlights 9 of the most impactful, newsworthy, and novel studies published in the journal over the past year:
Learn more: ja.ma/4rN3jM1

Chosen by JAMA editors, the inaugural roundup highlights 9 of the most impactful, newsworthy, and novel studies published in the journal over the past year:
Learn more: ja.ma/4rN3jM1
…the future of germline testing is clear now: generic first tier testing by long-read genome sequencing!

…the future of germline testing is clear now: generic first tier testing by long-read genome sequencing!
www.nature.com/articles/s41...

www.nature.com/articles/s41...

@hhu.de @crg.eu @embl.org @impvienna.bsky.social
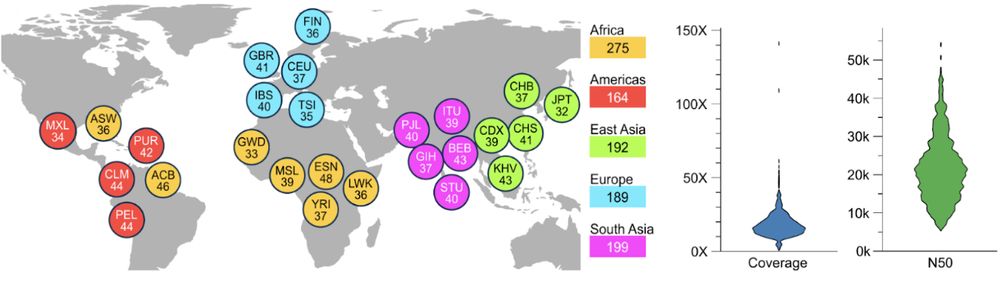
@hhu.de @crg.eu @embl.org @impvienna.bsky.social
Code: github.com/Platinum-Ped...
Data: github.com/Platinum-Ped...




Code: github.com/Platinum-Ped...
Data: github.com/Platinum-Ped...
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
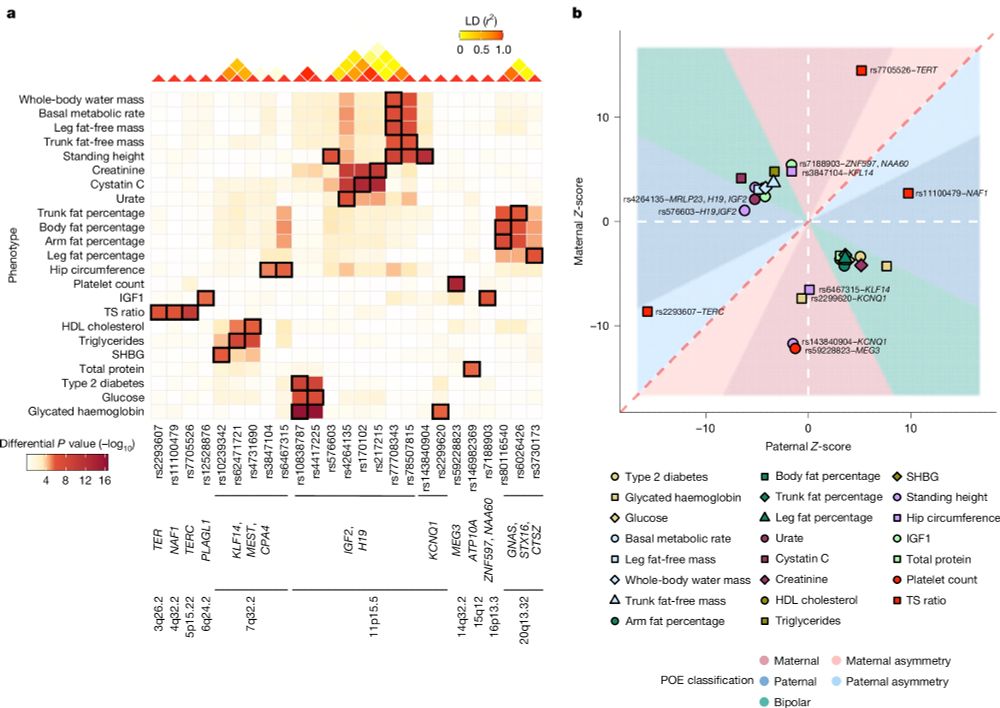
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
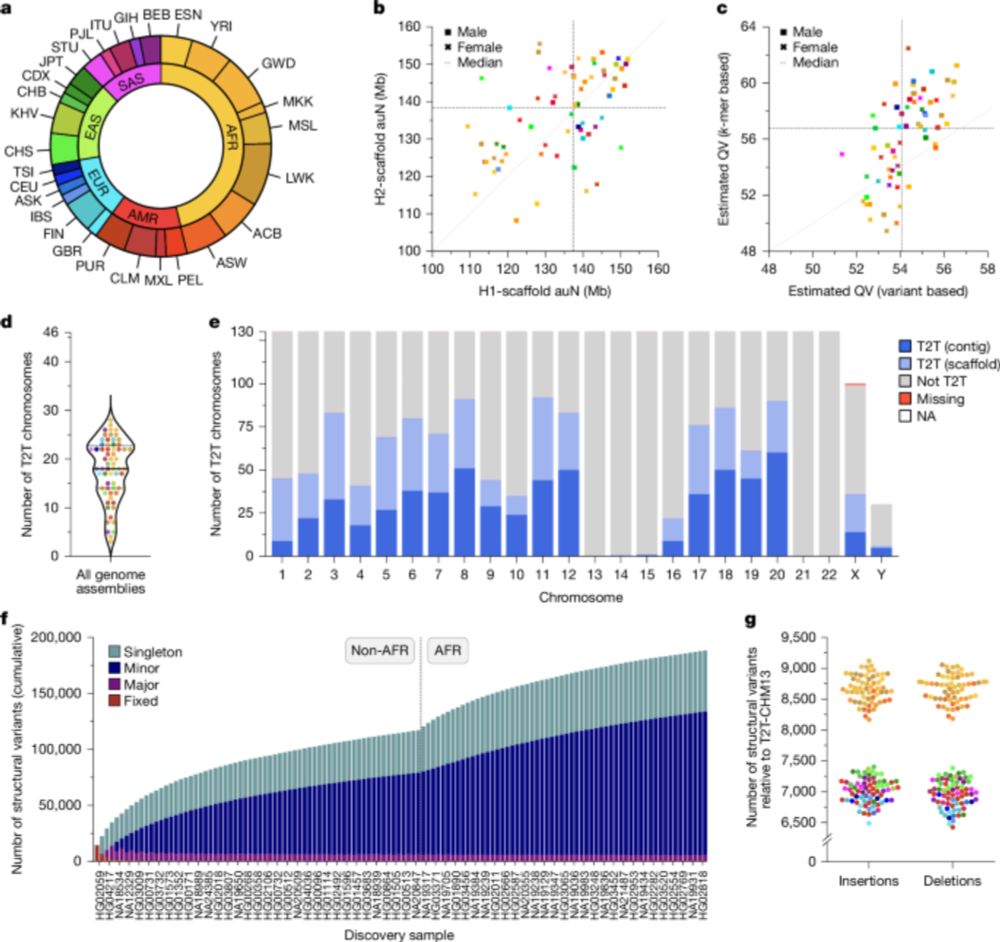
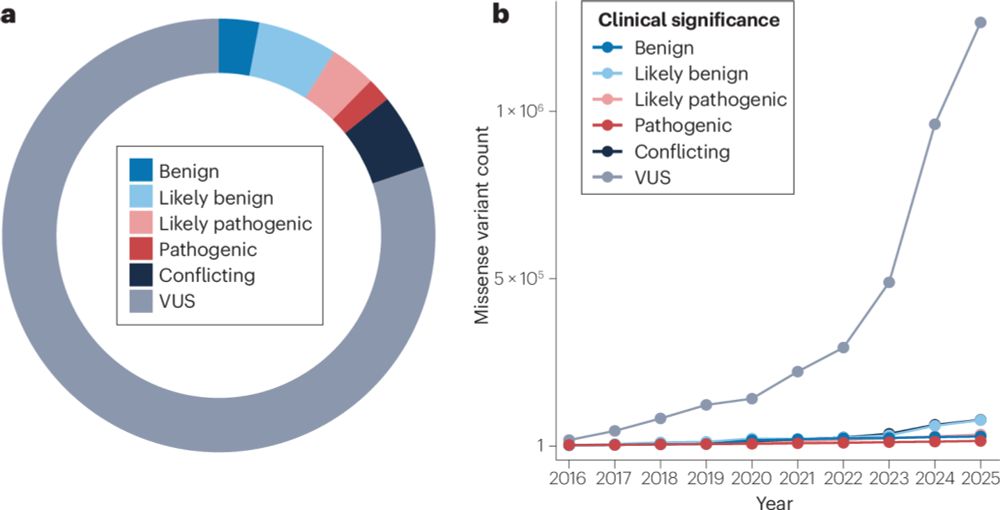
www.nature.com/articles/s41...
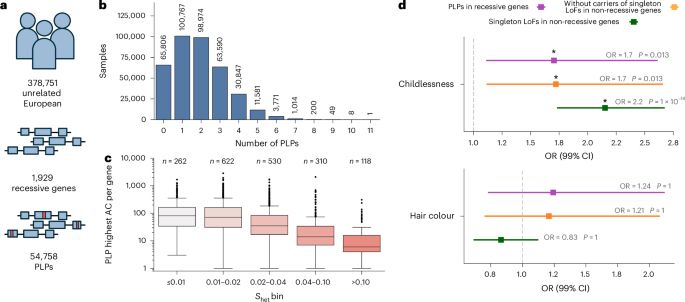
www.nature.com/articles/s41...

But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
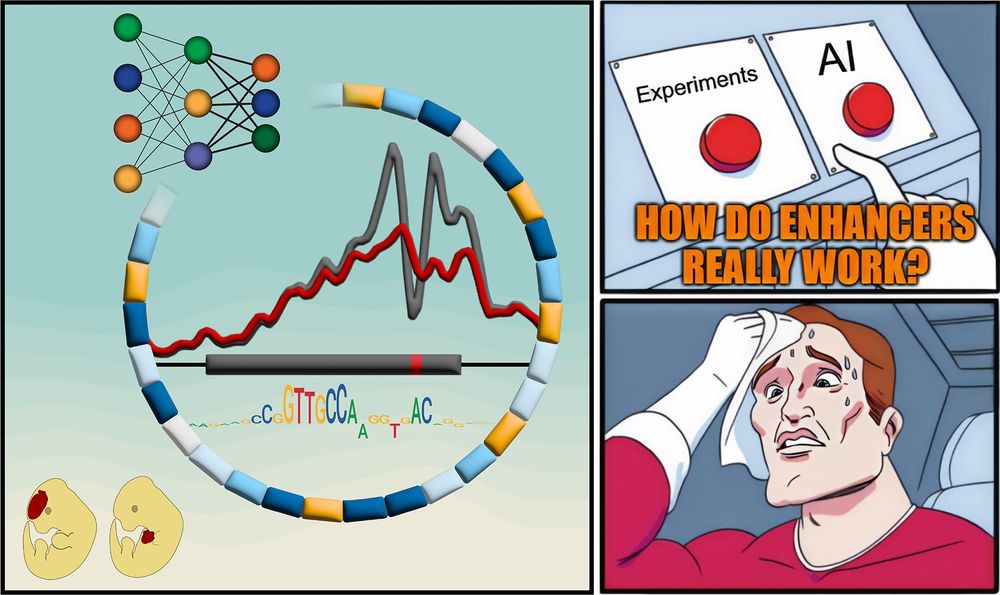
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22

Paying a lot of attention to the entire event will pay off!

Paying a lot of attention to the entire event will pay off!

…I could talk for hours about the evolution and tremendous progress made for sequence and genomics!
@eshg.bsky.social @eshgyoung.bsky.social #bertinoro2025 #eshg2025
…I could talk for hours about the evolution and tremendous progress made for sequence and genomics!
@eshg.bsky.social @eshgyoung.bsky.social #bertinoro2025 #eshg2025
tinyurl.com/Genome-Res-3...

🗳️Voting opens 28 April and runs until 11 May
🧬Your voice matters – take part!
@eshg.bsky.social #elections

🗳️Voting opens 28 April and runs until 11 May
🧬Your voice matters – take part!
@eshg.bsky.social #elections
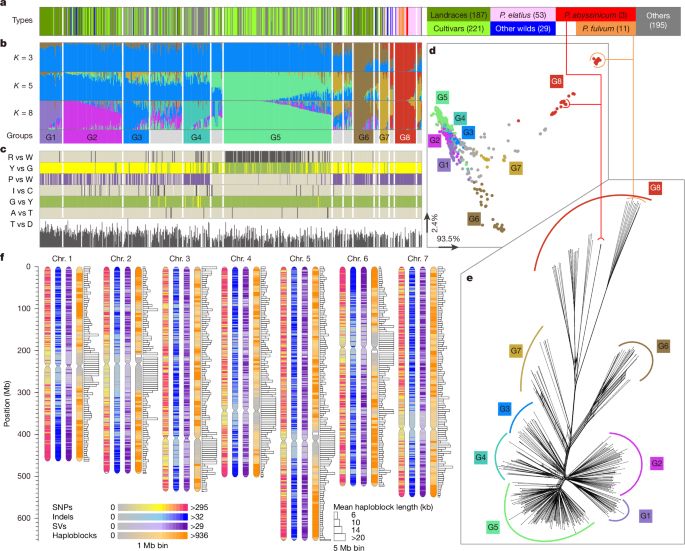
www.nature.com/articles/s41...

www.nature.com/articles/s41...
159 years after Mendel published his work, this is a real delight (especially for a geneticist)
🧪
@nature.com
www.nature.com/articles/s41...
159 years after Mendel published his work, this is a real delight (especially for a geneticist)
🧪
@nature.com
www.nature.com/articles/s41...

www.nature.com/articles/s41...
A thread (1/n)

www.nature.com/articles/s41...
A thread (1/n)
www.pacb.com/press_releas...

www.pacb.com/press_releas...


