The SPRQ-Nx chemistry, now in beta access, delivers complete genomes for under $300 at scale.
View the press release: bit.ly/4o06eyH

The SPRQ-Nx chemistry, now in beta access, delivers complete genomes for under $300 at scale.
View the press release: bit.ly/4o06eyH

Moar Rust plz!
github.com/PacificBiosc...
Moar Rust plz!
github.com/PacificBiosc...
@nature.com @uwgenome.bsky.social @eichlerlab.bsky.social @uwmedicine.bsky.social @utah.edu @pacbio.bsky.social

@nature.com @uwgenome.bsky.social @eichlerlab.bsky.social @uwmedicine.bsky.social @utah.edu @pacbio.bsky.social
www.nature.com/articles/s41...

www.nature.com/articles/s41...


github.com/PacificBiosc...
github.com/PacificBiosc...
Pre-print: doi.org/10.1101/2024...
Repo: github.com/PacificBiosc...

Pre-print: doi.org/10.1101/2024...
Repo: github.com/PacificBiosc...
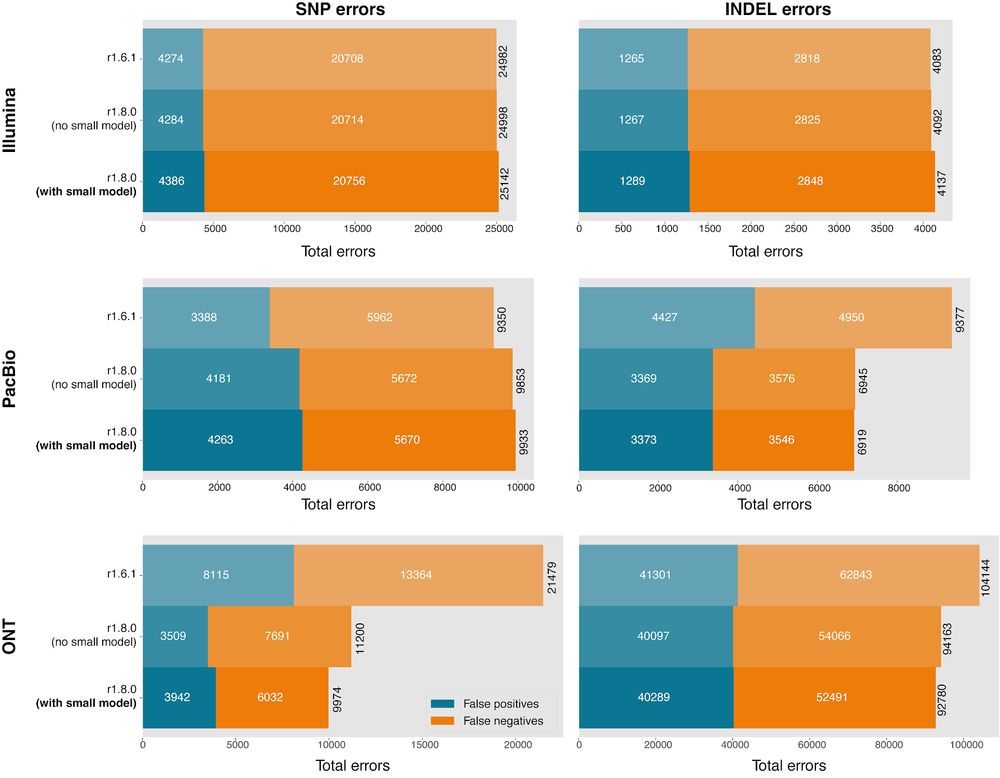
(github.com/google/deepv...)
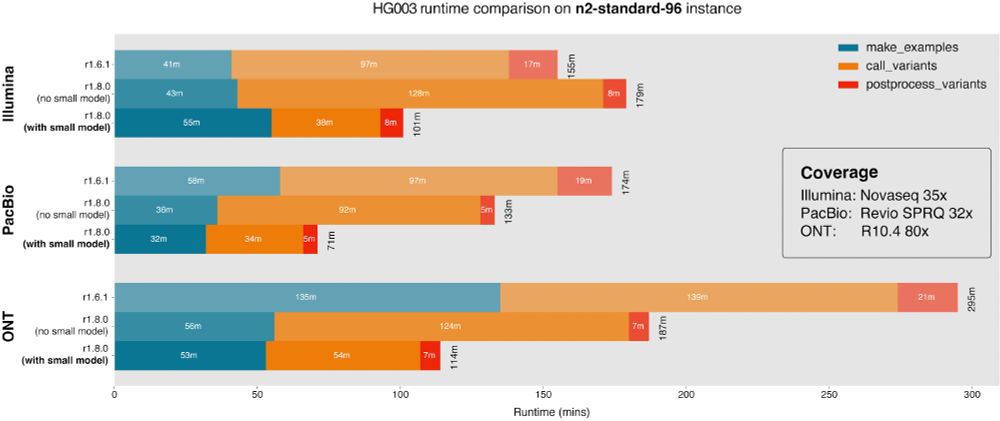
(github.com/google/deepv...)
Super excited about all the new work and special thanks to all the authors! Big thanks to Hillary @ahoischen.bsky.social @anaconesa.bsky.social

Super excited about all the new work and special thanks to all the authors! Big thanks to Hillary @ahoischen.bsky.social @anaconesa.bsky.social
Special thanks to Justin Zook from GIAB!

Special thanks to Justin Zook from GIAB!
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...