


Tiny, abundant, and anything but boring, phages shape microbial communities, move genes around, and can tip the balance in all kinds of ecosystems.
The catch: to know a #phage only you must know its host. ProxiMeta Hi-C technology gives us that link.

Tiny, abundant, and anything but boring, phages shape microbial communities, move genes around, and can tip the balance in all kinds of ecosystems.
The catch: to know a #phage only you must know its host. ProxiMeta Hi-C technology gives us that link.
na2.hubs.ly/H01HQ2Q0

na2.hubs.ly/H01HQ2Q0


Researchers assembled a chromosome-scale #genome for Deroceras laeve using Proximo Hi-C and @pacbio.bsky.social - making it a useful model for study of #genomics, #evolution, and slimyness... Congrats to Alfredo Varela-Echavarría & team!
🔗 academic.oup.com/g3journal/ad...

Researchers assembled a chromosome-scale #genome for Deroceras laeve using Proximo Hi-C and @pacbio.bsky.social - making it a useful model for study of #genomics, #evolution, and slimyness... Congrats to Alfredo Varela-Echavarría & team!
🔗 academic.oup.com/g3journal/ad...
Godspeed @phasegenomics.bsky.social
tinyurl.com/yaubcrpt

Godspeed @phasegenomics.bsky.social
tinyurl.com/yaubcrpt



Phase Genomics’ GPM works on #FFPE and mosaic samples. It’s just one assay, no metaphase prep, and no tradeoff in resolution. A new preprint shows how: www.medrxiv.org/content/10.1...
Phase Genomics’ GPM works on #FFPE and mosaic samples. It’s just one assay, no metaphase prep, and no tradeoff in resolution. A new preprint shows how: www.medrxiv.org/content/10.1...
🧬 130 haplotypes, many T2T
🧬 Closed 92% of assembly gaps
🧬 1,852 complex SVs
🧬 1,246 centromeres
www.nature.com/articles/s41...

📄 hubs.la/Q03ypPmb0
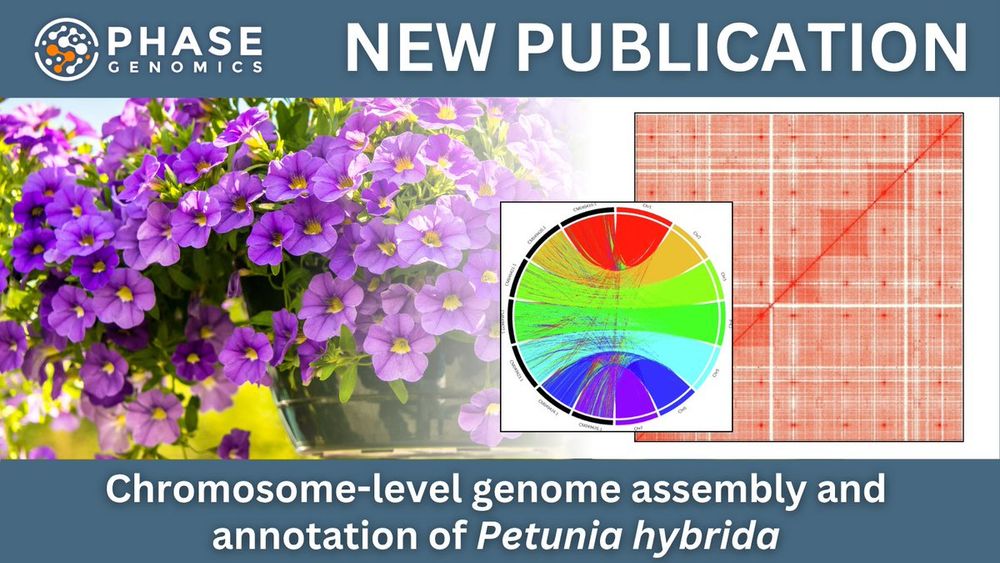
📄 hubs.la/Q03ypPmb0

@pacbio.bsky.social for the win! 🧬
🔗 hubs.la/Q03tZvZD0
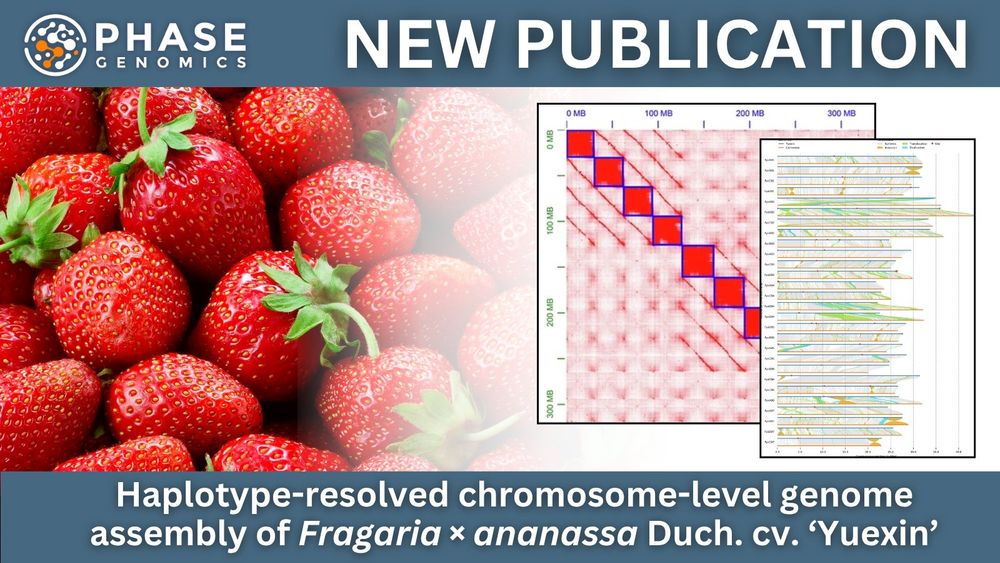
@pacbio.bsky.social for the win! 🧬
🔗 hubs.la/Q03tZvZD0
— Jon Chee, CEO @ Excedr
In the finale of The Biotech Startups Podcast, Jon and @ivanliachko.bsky.social unpack how smart science powers Phase Genomics' impact in cancer research, metagenomics & more👇
youtu.be/fqL6ZvS2sNQ?...

— Jon Chee, CEO @ Excedr
In the finale of The Biotech Startups Podcast, Jon and @ivanliachko.bsky.social unpack how smart science powers Phase Genomics' impact in cancer research, metagenomics & more👇
youtu.be/fqL6ZvS2sNQ?...
🔗https://hubs.la/Q03tZ1Jf0
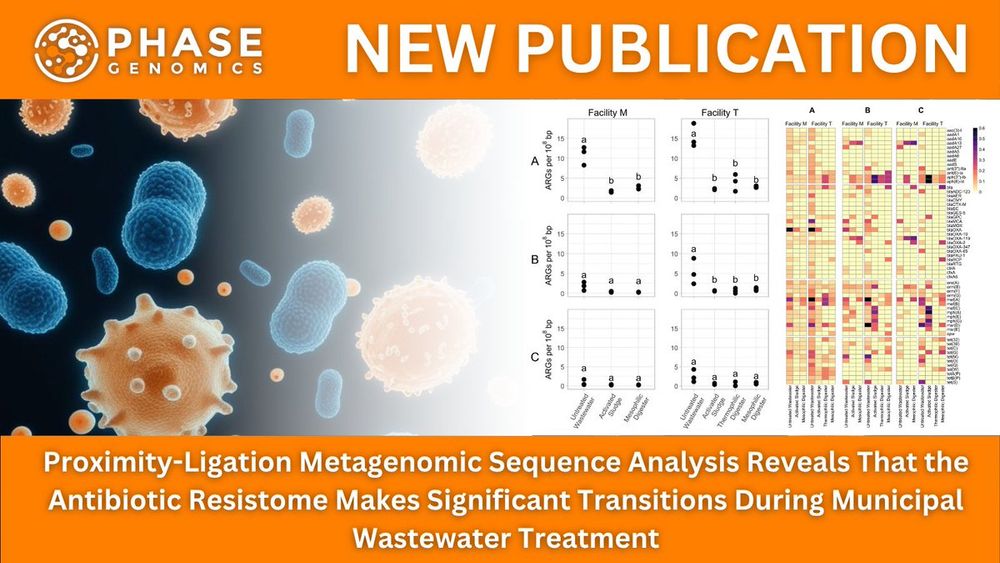
🔗https://hubs.la/Q03tZ1Jf0
🔗 Full study: hubs.la/Q03tZ6Ch0
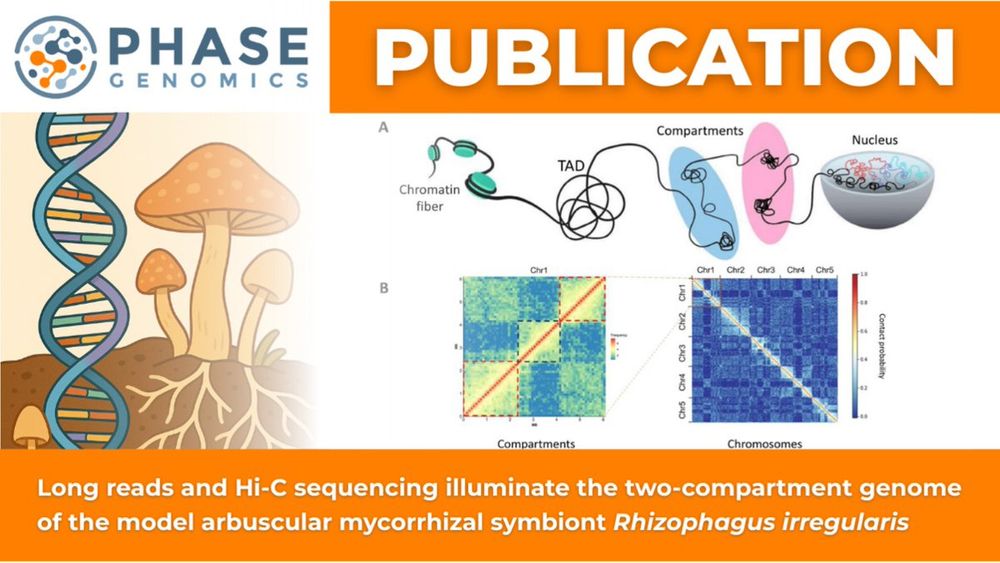
🔗 Full study: hubs.la/Q03tZ6Ch0
Hear how a postdoc aha moment became a platform accelerating #microbiome research, tackling #AMR & more on the Biotech Startups Podcast: www.youtube.com/watch?v=U2UG...

Hear how a postdoc aha moment became a platform accelerating #microbiome research, tackling #AMR & more on the Biotech Startups Podcast: www.youtube.com/watch?v=U2UG...
Researchers built #genomes of rough-seeded lupins - L. cosentinii & L. digitatus - revealing unique #gene expansions tied to stress resilience and seed traits. A howling good resource for legume breeding! #HiC
🔗 hubs.la/Q03v1F2c0

Researchers built #genomes of rough-seeded lupins - L. cosentinii & L. digitatus - revealing unique #gene expansions tied to stress resilience and seed traits. A howling good resource for legume breeding! #HiC
🔗 hubs.la/Q03v1F2c0



