
andrewcarroll.github.io/2026/02/09/f...
andrewcarroll.github.io/2026/02/09/f...
To learn more: blog.google/innovation-a...




To learn more: blog.google/innovation-a...
@ebpgenome.bsky.social. To support it Google.org has funded sequencing and open release of 13 genomes, with a $3M commit to sequence 150 more and develop methods to improve assembly finishing and other bottlenecks.
blog.google/innovation-a...

@ebpgenome.bsky.social. To support it Google.org has funded sequencing and open release of 13 genomes, with a $3M commit to sequence 150 more and develop methods to improve assembly finishing and other bottlenecks.
blog.google/innovation-a...

andrewcarroll.github.io/2025/12/23/t...
andrewcarroll.github.io/2025/12/23/t...
www.biorxiv.org/content/10.1...
#hoiho #conservation #genomics #birds #nzwildlife #endangered #wildlife #nature

www.biorxiv.org/content/10.1...
#hoiho #conservation #genomics #birds #nzwildlife #endangered #wildlife #nature



DV: Now train on HG002 T2T-Q100. Error reduction of 12% for Illumina and 30% for PacBio on this truth set. 25% faster. DeepTrio is 5x faster (20h -> 4h).
DS: New models FFPE_TUMOR_ONLY for {WGS, WES}. Much improved WGS models.
github.com/google/deepv...

DV: Now train on HG002 T2T-Q100. Error reduction of 12% for Illumina and 30% for PacBio on this truth set. 25% faster. DeepTrio is 5x faster (20h -> 4h).
DS: New models FFPE_TUMOR_ONLY for {WGS, WES}. Much improved WGS models.
github.com/google/deepv...
"We have fought and died alongside you....During your darkest hours...we were always there. Standing with you, grieving with you, the American people....Canadians are a little perplexed as to why our closest friends and neighbors are choosing to target us."

"We have fought and died alongside you....During your darkest hours...we were always there. Standing with you, grieving with you, the American people....Canadians are a little perplexed as to why our closest friends and neighbors are choosing to target us."
(github.com/google/deepv...)
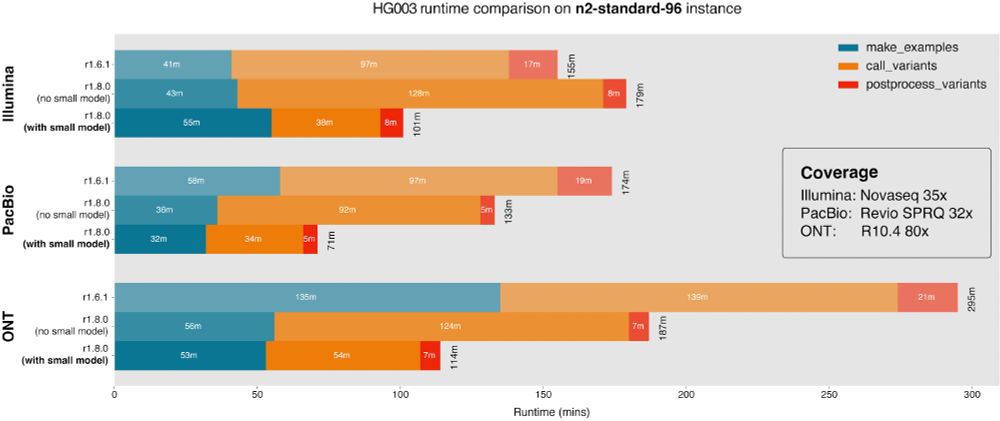
(github.com/google/deepv...)

Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...

Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...
(github.com/google/deeps...)

(github.com/google/deeps...)
I tried the subcellular locations in HPA but the membrane annotation is mostly fully intracellular proteins.
🧪🧬🖥️🔬
I tried the subcellular locations in HPA but the membrane annotation is mostly fully intracellular proteins.
🧪🧬🖥️🔬

