And community contributions from Sam Yadav and Seraj Ahmad at Roche improving the code for custom model training
And community contributions from Sam Yadav and Seraj Ahmad at Roche improving the code for custom model training
Thanks to student researcher contributions from Farica Zhuang and Mobin Asri.
DeepSomatic release page:
github.com/google/deeps...

Thanks to student researcher contributions from Farica Zhuang and Mobin Asri.
DeepSomatic release page:
github.com/google/deeps...
For running on GPU, I am not sure if you've seen this - github.com/google/deepv...
Which requires a little more configuration, but can let you better manage CPU-GPU tradeoffs. Definitely expert use.
For running on GPU, I am not sure if you've seen this - github.com/google/deepv...
Which requires a little more configuration, but can let you better manage CPU-GPU tradeoffs. Definitely expert use.
It's possible that pangenome-aware models will take more memory, and we do observe more memory per thread used for that. Definitely not lower than 16GB.
It's possible that pangenome-aware models will take more memory, and we do observe more memory per thread used for that. Definitely not lower than 16GB.
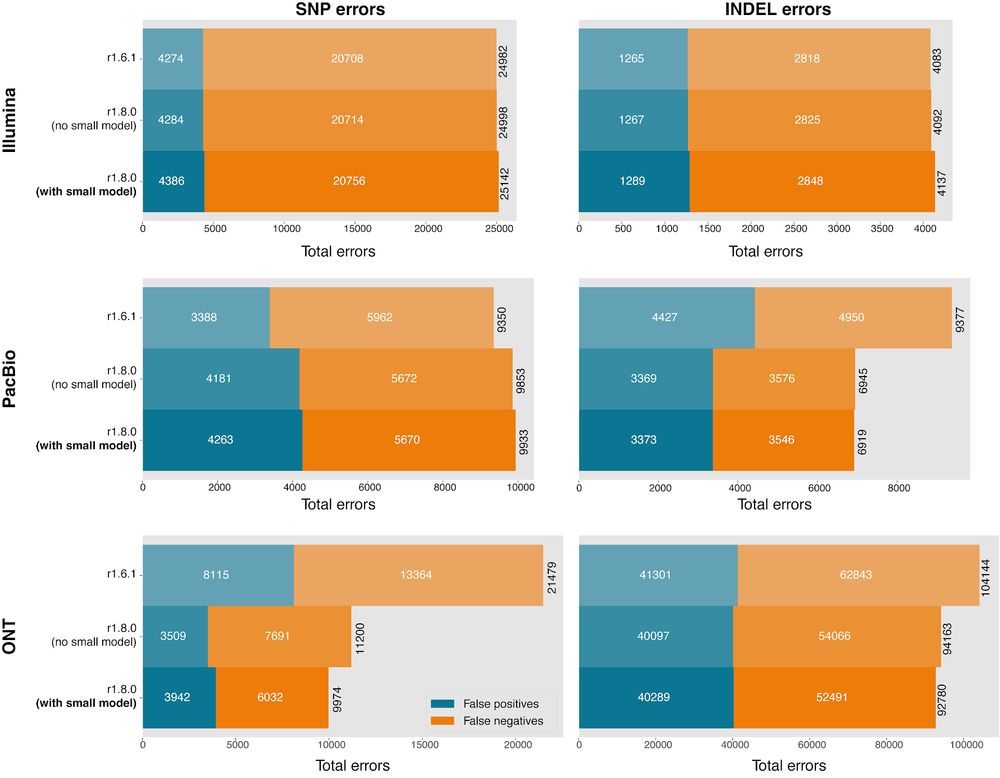
Major contributions from Pi-Chuan Chang, Daniel Cook, Alexey Kolesnikov
Google 20%ers: Will Kwan, Pauline Sho, Lucas Brambrink, Mo Samman, Atilla Kiraly
UCSC for vg: @benedictpaten.bsky.social, Shloka Negi, Jimin Park, Mobin Asri
Pacbio: Billy Rowell, Nathaniel Echols
Major contributions from Pi-Chuan Chang, Daniel Cook, Alexey Kolesnikov
Google 20%ers: Will Kwan, Pauline Sho, Lucas Brambrink, Mo Samman, Atilla Kiraly
UCSC for vg: @benedictpaten.bsky.social, Shloka Negi, Jimin Park, Mobin Asri
Pacbio: Billy Rowell, Nathaniel Echols
T7: github.com/google/deepv...
G400: github.com/google/deepv...
For now, these are stand alone models. We'll likely consider whether we can jointly include these in the broad WGS model later.
T7: github.com/google/deepv...
G400: github.com/google/deepv...
For now, these are stand alone models. We'll likely consider whether we can jointly include these in the broad WGS model later.
github.com/google/deepv...
github.com/google/deepv...
github.com/google/deepv...
Shows a step by step process, with Docker images for how to map to a Pangenome reference w/ vg and calls w/ DeepVariant. Final calls are more accurate and in GRCh38 coordinates. Thanks to the UCSC team for co-development
github.com/google/deepv...
Shows a step by step process, with Docker images for how to map to a Pangenome reference w/ vg and calls w/ DeepVariant. Final calls are more accurate and in GRCh38 coordinates. Thanks to the UCSC team for co-development
(github.com/KolmogorovLa...)
Thanks to Khi Pin Chua and Billy Rowell from PacBio

(github.com/KolmogorovLa...)
Thanks to Khi Pin Chua and Billy Rowell from PacBio

