jobs.uri.edu/postings/15960

jobs.uri.edu/postings/15960
This was a wonderful collaboration with Sam Vohsen, Eslam Osman, Mandy Joye, Matt Saxton, grubervodicka.bsky.social, @ibaums.bsky.social

This was a wonderful collaboration with Sam Vohsen, Eslam Osman, Mandy Joye, Matt Saxton, grubervodicka.bsky.social, @ibaums.bsky.social

pmc.ncbi.nlm.nih.gov/articles/PMC...
You can read our news and views here: www.pnas.org/doi/10.1073/...
pmc.ncbi.nlm.nih.gov/articles/PMC...
You can read our news and views here: www.pnas.org/doi/10.1073/...
apply.interfolio.com/177547
apply.interfolio.com/177547

this project was led by @omernadel.bsky.social and is a joint work between the labs of @bejalab.bsky.social, Debbie Lindell and Oded Kleifeld from @biologytechnion.bsky.social

this project was led by @omernadel.bsky.social and is a joint work between the labs of @bejalab.bsky.social, Debbie Lindell and Oded Kleifeld from @biologytechnion.bsky.social
Deadline: Dec. 2, 2 p.m. ET.
Share your science → asm.social/2Gv

Deadline: Dec. 2, 2 p.m. ET.
Share your science → asm.social/2Gv
Bringing together scientists from diverse fields to foster new discoveries in condensate biology.
Submit your abstract by 10 Feb: s.embl.org/ees26-08-bl
📅 19 – 22 May 2026

Bringing together scientists from diverse fields to foster new discoveries in condensate biology.
Submit your abstract by 10 Feb: s.embl.org/ees26-08-bl
📅 19 – 22 May 2026
Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...

Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...
cluster-ia-enact.ai/appels-a-pro...
Deadline: Jan 7, 2026

cluster-ia-enact.ai/appels-a-pro...
Deadline: Jan 7, 2026

cluster-ia-enact.ai/appels-a-pro...
Deadline: Jan 7, 2026
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
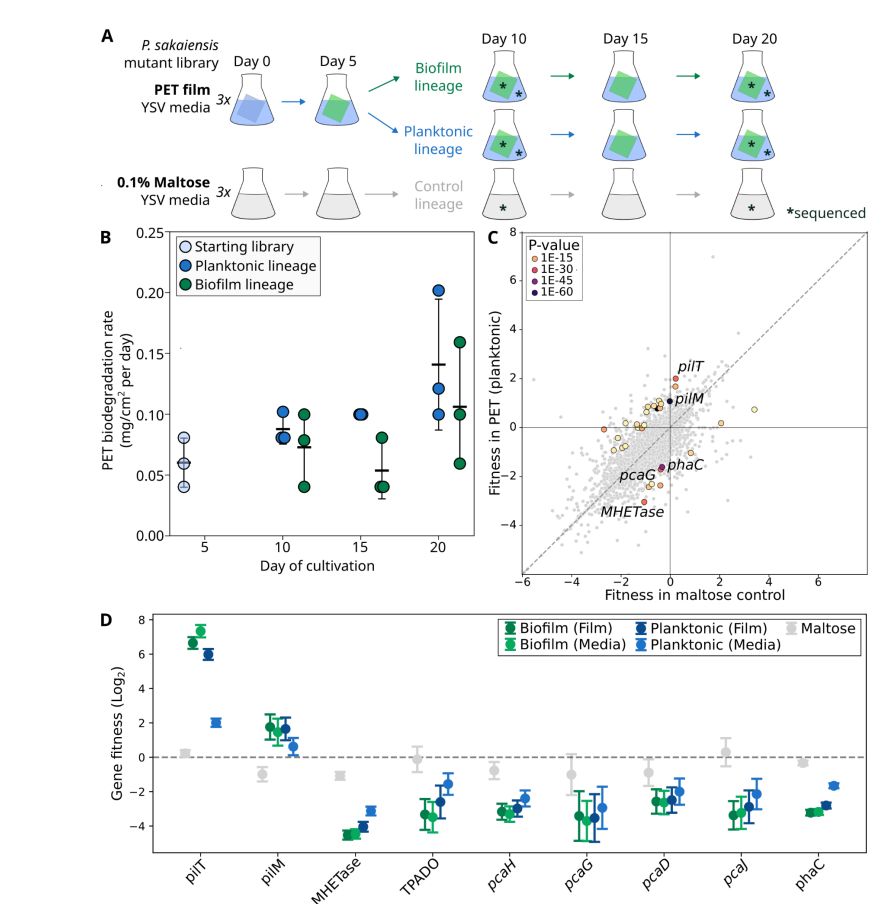
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.

Thanks to amazing speakers for helping us realise this!
Join the brand-new EMBO | EMBL Symposium #EESCollectivity and explore how collective behaviours arise from fundamental principles across biological systems 🧬🦠🐒
💻 s.embl.org/ees26-01-bl
✒️ Submit your abstract by 18 Nov

Thanks to amazing speakers for helping us realise this!

tinyurl.com/ch3damp
We show how molecular byproducts released during virus-induced cell exploitation are used as signals to trigger host immunity
Revealed by the amazing Ilya Osterman. See his thread below👇

tinyurl.com/ch3damp
We show how molecular byproducts released during virus-induced cell exploitation are used as signals to trigger host immunity
Revealed by the amazing Ilya Osterman. See his thread below👇

🔗 www.embl.org/news/science...

🔗 www.embl.org/news/science...

Very cool paper and method. I was looking forward to this coming out of preprint (short thread below on why)
www.nature.com/articles/s41...

Very cool paper and method. I was looking forward to this coming out of preprint (short thread below on why)
www.nature.com/articles/s41...
apply.interfolio.com/176402
apply.interfolio.com/176402
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

Henipaviruses, like Nipah and Hendra, package their genomes inside helical shells built by thousands of nucleoproteins. These nucleocapsids are essential to protect the viral RNA, but how do they ever let the polymerase in to read the sequence?
👇

Henipaviruses, like Nipah and Hendra, package their genomes inside helical shells built by thousands of nucleoproteins. These nucleocapsids are essential to protect the viral RNA, but how do they ever let the polymerase in to read the sequence?
👇

