Using metagenomic, single-bacterial, and biochemical methods we provide insights into uncultivated bacterial symbionts as rich sources of bioactive natural products in marine sponges.
#DrugDiscovery #NaturalProducts

Using metagenomic, single-bacterial, and biochemical methods we provide insights into uncultivated bacterial symbionts as rich sources of bioactive natural products in marine sponges.
#DrugDiscovery #NaturalProducts
Pieter C Dorrestein & Nicole E Avalon
#naturalproducts #secmet #metabolomics
www.sciencedirect.com/science/arti...

Pieter C Dorrestein & Nicole E Avalon
#naturalproducts #secmet #metabolomics
www.sciencedirect.com/science/arti...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
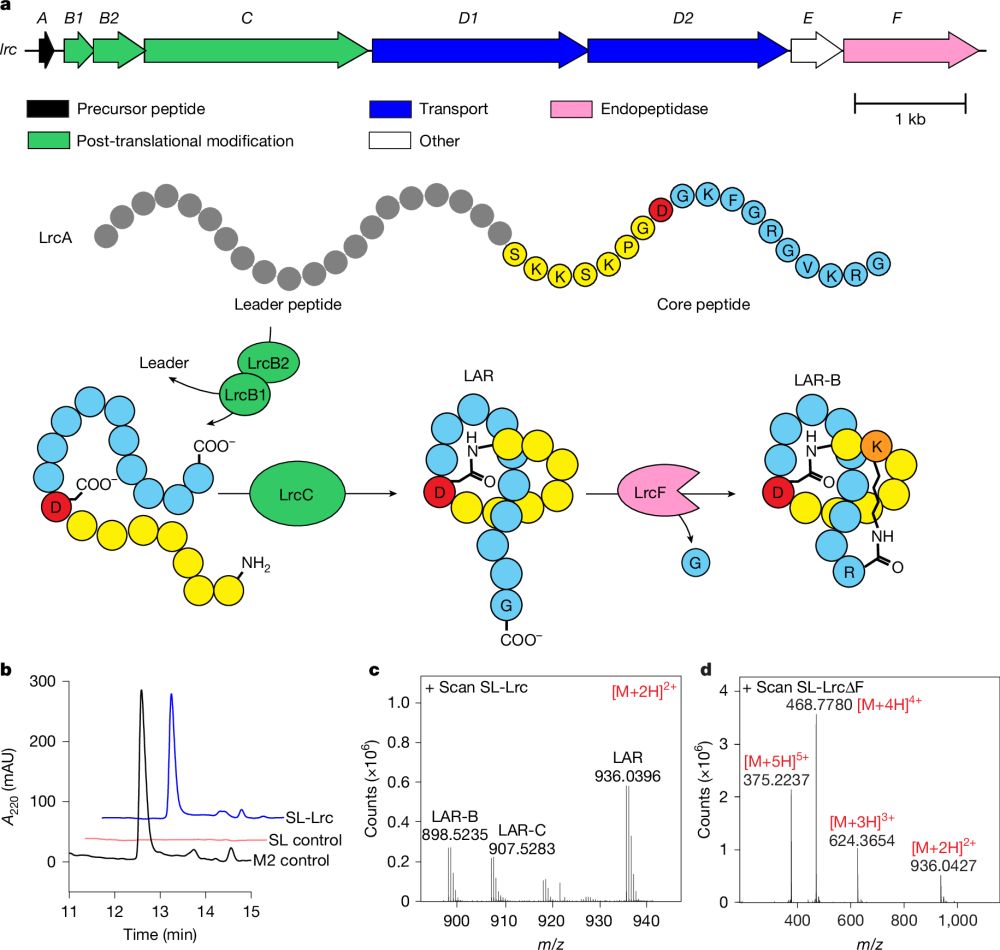
#secmet #lassopeptides #syntheticbiology

#secmet #lassopeptides #syntheticbiology

Find it in full below👇

#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...

#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...
And check out the Secondary Metabolism feed. Lots of awesome science here on Bluesky!
bsky.app/profile/did:...
And check out the Secondary Metabolism feed. Lots of awesome science here on Bluesky!
bsky.app/profile/did:...

doi.org/10.1101/2025...

doi.org/10.1101/2025...
Our team’s first conference ended on a high note — 2nd prize for best poster! 🥈🥳
Thanks to the organizers @niedermeyer-lab.bsky.social for a fantastic meeting. 🙏

Our team’s first conference ended on a high note — 2nd prize for best poster! 🥈🥳
Thanks to the organizers @niedermeyer-lab.bsky.social for a fantastic meeting. 🙏
We just published the paper Biosynthesis of biologically active terpenoids in the mint family (Lamiaceae) in Natural Products Reports.
Thanks to everyone involved! :)

We just published the paper Biosynthesis of biologically active terpenoids in the mint family (Lamiaceae) in Natural Products Reports.
Thanks to everyone involved! :)
pubs.acs.org/doi/10.1021/...
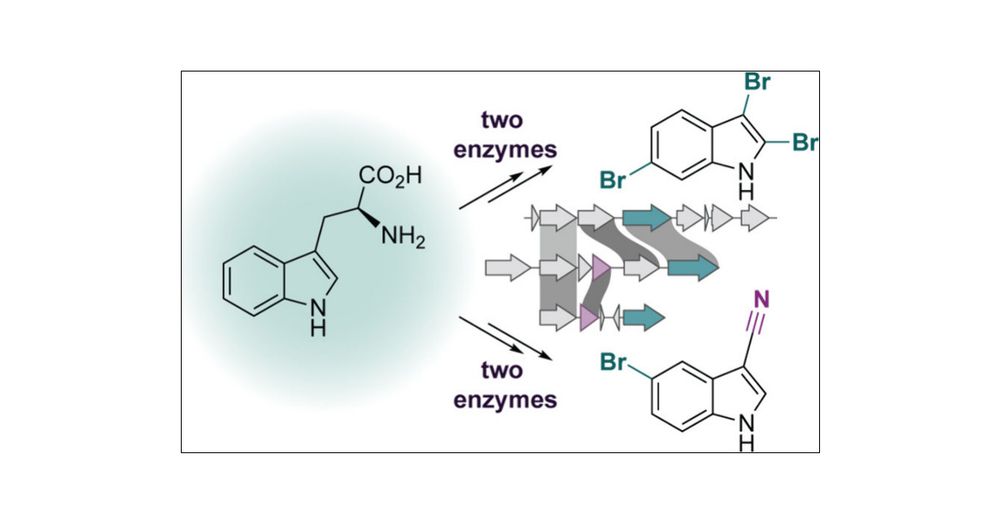
pubs.acs.org/doi/10.1021/...
Fantastic team effort with the Pucker group @bpucker.bsky.social @puckerlab.bsky.social at @unibonn.bsky.social and Heretsch, Herde and Witte groups at @unihannover.bsky.social. Funded by @dfg.de
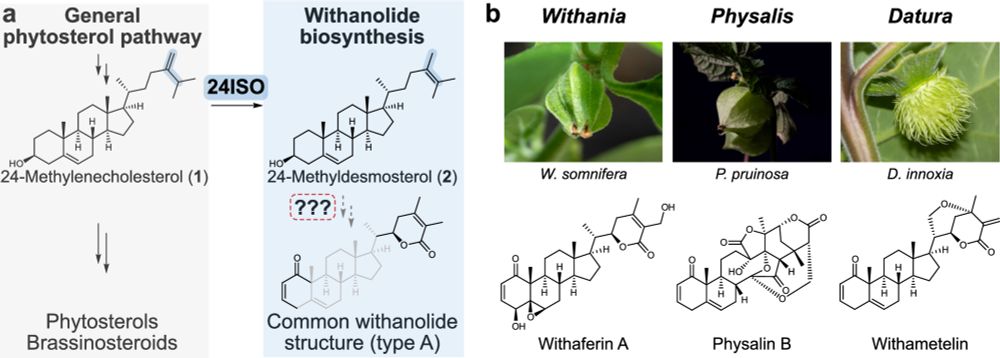
Fantastic team effort with the Pucker group @bpucker.bsky.social @puckerlab.bsky.social at @unibonn.bsky.social and Heretsch, Herde and Witte groups at @unihannover.bsky.social. Funded by @dfg.de
www.nature.com/articles/s41...
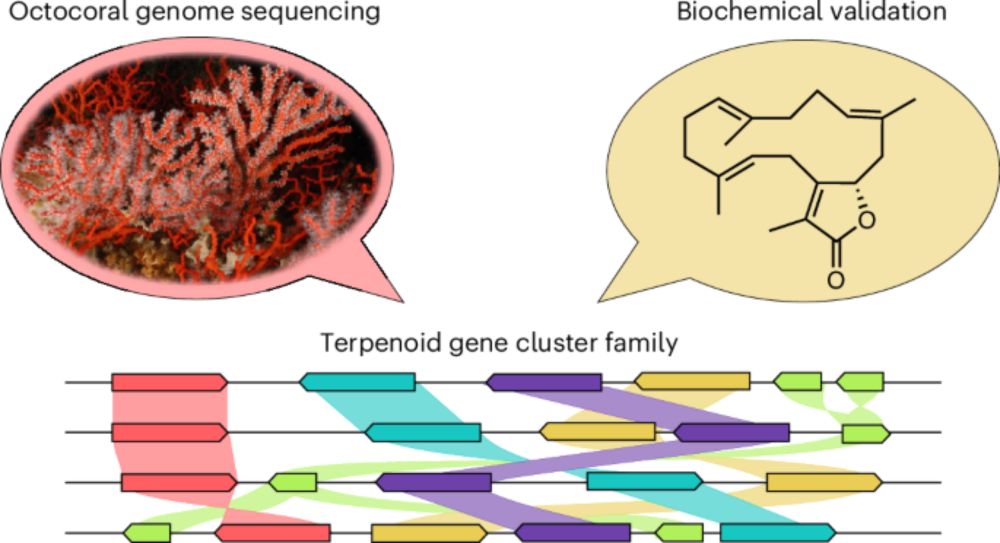
www.nature.com/articles/s41...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
→ 800+ MAGs
→ 11,000+ BGCs
→ and still nowhere near saturation.
Soil is wild.
Preprint here: www.biorxiv.org/content/10.1...
#microbiome #metagenomics #BGCs #naturalproducts #secmet

→ 800+ MAGs
→ 11,000+ BGCs
→ and still nowhere near saturation.
Soil is wild.
Preprint here: www.biorxiv.org/content/10.1...
#microbiome #metagenomics #BGCs #naturalproducts #secmet
-->Cutting-edge research in an open, collaborative environment? Check out the call and join one of Germany’s leading research universities.
#AcademicJobs #FacultyPosition
www.nature.com/naturecareer...

-->Cutting-edge research in an open, collaborative environment? Check out the call and join one of Germany’s leading research universities.
#AcademicJobs #FacultyPosition
www.nature.com/naturecareer...