Marnix Medema
@marnixmedema.bsky.social
Professor of Bioinformatics at Wageningen University, Visiting professor at Leiden University. Natural product discovery, microbiomes, method development.
Reposted by Marnix Medema
Happy to share our newest manuscript about the discovery and hererologous expression of metanodin, a new lassopeptide with unprecedented structural features directly from soil metagenomes. pubs.acs.org/doi/full/10....
#secmet #lassopeptides #syntheticbiology
#secmet #lassopeptides #syntheticbiology

Discovery and Heterologous Expression of the Soil Metagenome-Derived Lasso Peptide Metanodin with an Unprecedented Ring Structure
Culture-independent metagenomic approaches have proven to be effective tools for identifying previously hidden biosynthetic gene clusters (BGCs) encoding novel natural products with potential medical relevance. However, producing these compounds remains challenging as metagenomic BGCs often originate from organisms phylogenetically distant from available heterologous hosts. Lasso peptides, a subclass of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products, exhibit diverse bioactivities, yet no lasso peptide has previously been discovered directly from a metagenome. Here, we report the discovery and heterologous expression of the first soil metagenome-derived lasso peptide. Expression of its biosynthetic gene cluster in Escherichia coli, followed by mass spectrometry analysis, strongly supported the predicted amino acid sequence and lasso structure of the peptide. Notably, this lasso peptide is the first to feature asparagine as the ring-forming residue at position one. Taxonomic analysis of the corresponding BGC identified an uncultivated member of the Steroidobacterales family (Gammaproteobacteria) as the closest known relative of the potential native host. These findings underscore the potential of metagenomic genome mining to reveal structurally novel RiPPs and to expand our understanding of the natural diversity of lasso peptides.
pubs.acs.org
October 28, 2025 at 2:39 PM
Happy to share our newest manuscript about the discovery and hererologous expression of metanodin, a new lassopeptide with unprecedented structural features directly from soil metagenomes. pubs.acs.org/doi/full/10....
#secmet #lassopeptides #syntheticbiology
#secmet #lassopeptides #syntheticbiology
Kudos to @elenadelpup.bsky.social for this significant update to plantiSMASH! Try it out now...
🌱plantiSMASH 2.0💥 is now live!
I’m very excited to share that the 2.0 version of plantiSMASH is now available online and as a preprint on bioRxiv 📄https://lnkd.in/dN4F56pf
🌱 What’s new in plantiSMASH 2.0? ⬇️
I’m very excited to share that the 2.0 version of plantiSMASH is now available online and as a preprint on bioRxiv 📄https://lnkd.in/dN4F56pf
🌱 What’s new in plantiSMASH 2.0? ⬇️
plantiSMASH 2.0: improvements to detection, annotation, and prioritization of plant biosynthetic gene clusters https://www.biorxiv.org/content/10.1101/2025.10.28.683968v1
October 29, 2025 at 2:04 PM
Kudos to @elenadelpup.bsky.social for this significant update to plantiSMASH! Try it out now...
Reposted by Marnix Medema
Polyunsaturated fatty acid (PUFA) synthase enzymes turn out to be far more versatile than we ever imagined! In our new preprint, we reveal a largely untapped biosynthetic space where they team up with PKSs and NRPSs to create new types of bioactive amphiphilic metabolites.
👉http://bit.ly/4hzUUak
👉http://bit.ly/4hzUUak

Charting the biosynthetic landscape of hybrid polyketide-nonribosomal peptide-specialized lipids
Polyunsaturated fatty acid (PUFA) synthase-like enzymes are best known for their role in membrane lipid biosynthesis in marine bacteria, but have also been repurposed for the assembly of specialized l...
bit.ly
October 27, 2025 at 9:56 AM
Polyunsaturated fatty acid (PUFA) synthase enzymes turn out to be far more versatile than we ever imagined! In our new preprint, we reveal a largely untapped biosynthetic space where they team up with PKSs and NRPSs to create new types of bioactive amphiphilic metabolites.
👉http://bit.ly/4hzUUak
👉http://bit.ly/4hzUUak
Reposted by Marnix Medema
Working on tailoring enzymes and want to learn more about the MITE database? We are organizing a hands-on training session on how use the DB and create new entries: October 30th and November 4th 10-11 am Amsterdam time. Sign up here: forms.gle/KHPqe9XV1RRg... See you there! #secmet #natprod

MITE training session registration form
Registration form for mailing list
forms.gle
October 24, 2025 at 1:37 PM
Working on tailoring enzymes and want to learn more about the MITE database? We are organizing a hands-on training session on how use the DB and create new entries: October 30th and November 4th 10-11 am Amsterdam time. Sign up here: forms.gle/KHPqe9XV1RRg... See you there! #secmet #natprod
Join this session if you want to learn more about MITE and contributing to it!
Working on tailoring enzymes and want to learn more about the MITE database? We are organizing a hands-on training session on how use the DB and create new entries: October 30th and November 4th 10-11 am Amsterdam time. Sign up here: forms.gle/KHPqe9XV1RRg... See you there! #secmet #natprod

MITE training session registration form
Registration form for mailing list
forms.gle
October 24, 2025 at 3:15 PM
Join this session if you want to learn more about MITE and contributing to it!
Reposted by Marnix Medema
🧬 Share your science at Natural Products in the 21st Century (NP21C) — 9–11 Feb 2026, Leuven
Join researchers exploring genomics, metabolomics & microbial natural products!
www.conferencemanager.dk/naturalprodu...
#NP21C #secmet
@tilmweber.bsky.social @marnixmedema.bsky.social @kblin.bsky.social
Join researchers exploring genomics, metabolomics & microbial natural products!
www.conferencemanager.dk/naturalprodu...
#NP21C #secmet
@tilmweber.bsky.social @marnixmedema.bsky.social @kblin.bsky.social
Natural Products in the 21st Century Conference
www.conferencemanager.dk
October 23, 2025 at 8:28 PM
🧬 Share your science at Natural Products in the 21st Century (NP21C) — 9–11 Feb 2026, Leuven
Join researchers exploring genomics, metabolomics & microbial natural products!
www.conferencemanager.dk/naturalprodu...
#NP21C #secmet
@tilmweber.bsky.social @marnixmedema.bsky.social @kblin.bsky.social
Join researchers exploring genomics, metabolomics & microbial natural products!
www.conferencemanager.dk/naturalprodu...
#NP21C #secmet
@tilmweber.bsky.social @marnixmedema.bsky.social @kblin.bsky.social
Reposted by Marnix Medema
What a fantastic effort by @elenadelpup.bsky.social who brought together scientists today to discuss the implementation of knowledge graphs to describe and predict plant ☘️ biosynthesis pathways!
Watch our space 😊
Organized with support from @marnixmedema.bsky.social & myself 🙂
Watch our space 😊
Organized with support from @marnixmedema.bsky.social & myself 🙂

October 14, 2025 at 3:16 PM
What a fantastic effort by @elenadelpup.bsky.social who brought together scientists today to discuss the implementation of knowledge graphs to describe and predict plant ☘️ biosynthesis pathways!
Watch our space 😊
Organized with support from @marnixmedema.bsky.social & myself 🙂
Watch our space 😊
Organized with support from @marnixmedema.bsky.social & myself 🙂
Reposted by Marnix Medema
Welcome to #Wageningen @adafede.bsky.social, Pierre-Marie Allard, and Tito Damiani! 😎 Looking forward to the mini-symposium on multi-omics & knowledge graphs tomorrow organized by @elenadelpup.bsky.social with support from @marnixmedema.bsky.social & myself! 😊
#metabolomics #CompMetabolomics
#metabolomics #CompMetabolomics


October 13, 2025 at 8:33 PM
Welcome to #Wageningen @adafede.bsky.social, Pierre-Marie Allard, and Tito Damiani! 😎 Looking forward to the mini-symposium on multi-omics & knowledge graphs tomorrow organized by @elenadelpup.bsky.social with support from @marnixmedema.bsky.social & myself! 😊
#metabolomics #CompMetabolomics
#metabolomics #CompMetabolomics
Reposted by Marnix Medema
Great to see PhyloNaP out! Great tool, and it also connects to mite.bioinformatic.nl! #openscience #collaboration
September 27, 2025 at 11:21 AM
Great to see PhyloNaP out! Great tool, and it also connects to mite.bioinformatic.nl! #openscience #collaboration
Very important initiative! This could really help facilitate increasing data sharing as well as appropriate attribution of data creation.
New article on equitable reuse of public sequencing data, published in @natmicrobiol.nature.com!
Led by the Data reuse core team @lhug.bsky.social @environmicrobio.bsky.social Cristina Moraru, @geomicrosoares.bsky.social, @folker.bsky.social and with Anke Heyer and The Data Reuse Consotrium!
Led by the Data reuse core team @lhug.bsky.social @environmicrobio.bsky.social Cristina Moraru, @geomicrosoares.bsky.social, @folker.bsky.social and with Anke Heyer and The Data Reuse Consotrium!

September 29, 2025 at 6:13 PM
Very important initiative! This could really help facilitate increasing data sharing as well as appropriate attribution of data creation.
Reposted by Marnix Medema
Happy to share our newest preprint. PhyloNaP as a user friendly database of phylogeny for enzymes involved in natural product production and as public repository for well curated phylogenetic trees. Happy Tree Building!!!
#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...
#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...

PhyloNaP: a user-friendly database of Phylogeny for Natural Product-producing enzymes
Phylogenetic analysis is widely used to predict enzyme function, yet building annotated and reusable trees is labor-intensive and requires extensive knowledge about the specific enzymes. Existing reso...
www.biorxiv.org
September 27, 2025 at 9:32 AM
Happy to share our newest preprint. PhyloNaP as a user friendly database of phylogeny for enzymes involved in natural product production and as public repository for well curated phylogenetic trees. Happy Tree Building!!!
#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...
#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...
Awesome work, @mmzdouc.bsky.social !
Aaand it's out! Meet MITE - the natural product tailoring enzyme database, just published in @narjournal.bsky.social! MITE DB captures the substrate- and reaction-specificity of tailoring enzymes, allowing to capture this information in a human- and machine-readable way! doi.org/10.1093/nar/...
September 27, 2025 at 9:13 AM
Awesome work, @mmzdouc.bsky.social !
Reposted by Marnix Medema
Aaand it's out! Meet MITE - the natural product tailoring enzyme database, just published in @narjournal.bsky.social! MITE DB captures the substrate- and reaction-specificity of tailoring enzymes, allowing to capture this information in a human- and machine-readable way! doi.org/10.1093/nar/...
September 27, 2025 at 9:11 AM
Aaand it's out! Meet MITE - the natural product tailoring enzyme database, just published in @narjournal.bsky.social! MITE DB captures the substrate- and reaction-specificity of tailoring enzymes, allowing to capture this information in a human- and machine-readable way! doi.org/10.1093/nar/...
Congratulations to #YuzeLi and all co-authors with the publication of rhizoSMASH: a new tool to identify catabolic gene clusters involved in the metabolism of plant root exudates, thus driving rhizosphere colonization. 1/4
rdcu.be/eH8tr
rdcu.be/eH8tr

Predicting rhizosphere-competence-related catabolic gene clusters in plant-associated bacteria with rhizoSMASH
Nature Communications - Rhizosphere microbiomes are shaped by root exudation of diverse organic compounds. Here, the rhizoSMASH algorithm is introduced, which maps microbial genes involved in their...
rdcu.be
September 25, 2025 at 12:07 PM
Congratulations to #YuzeLi and all co-authors with the publication of rhizoSMASH: a new tool to identify catabolic gene clusters involved in the metabolism of plant root exudates, thus driving rhizosphere colonization. 1/4
rdcu.be/eH8tr
rdcu.be/eH8tr
New preprint out by #RobertKoetsier, the first of his PhD project, on assessing the use of cross-species coexpression analysis to identify primary and secondary metabolic interactions in microbiomes: www.biorxiv.org/content/10.1...

Using cross-species co-expression to predict metabolic interactions in microbiomes
In microbial ecosystems, metabolic interactions are key determinants of species’ relative abundance and activity. Given the immense number of possible interactions in microbial communities, their expe...
www.biorxiv.org
September 22, 2025 at 7:42 AM
New preprint out by #RobertKoetsier, the first of his PhD project, on assessing the use of cross-species coexpression analysis to identify primary and secondary metabolic interactions in microbiomes: www.biorxiv.org/content/10.1...
Reposted by Marnix Medema
Check out the new MITE database! Kudos to @mmzdouc.bsky.social for his leadership in getting this community effort off the ground. Hopefully we can add many more data points with everyone's input!
Into natural product biosynthesis & tailoring enzymes? Frustrated by the lack of a dedicated resource to explore their functions? Tired of endless literature searches for reaction info? Meet the MITE database, freely available at mite.bioinformatics.nl. Preprint: doi.org/10.26434/che... (1/8)

August 21, 2025 at 1:07 PM
Check out the new MITE database! Kudos to @mmzdouc.bsky.social for his leadership in getting this community effort off the ground. Hopefully we can add many more data points with everyone's input!
We are looking for a 3-year postdoc to work with Daniel Probst, Justin van der Hooft and myself on an exciting project involving federated learning and integrative omics for discovery of new antibiotics from natural products.
Apply here: www.wur.nl/en/vacancy/p...
Please share!
Apply here: www.wur.nl/en/vacancy/p...
Please share!
www.wur.nl
September 8, 2025 at 9:07 AM
We are looking for a 3-year postdoc to work with Daniel Probst, Justin van der Hooft and myself on an exciting project involving federated learning and integrative omics for discovery of new antibiotics from natural products.
Apply here: www.wur.nl/en/vacancy/p...
Please share!
Apply here: www.wur.nl/en/vacancy/p...
Please share!
Reposted by Marnix Medema
Now out as preprint: versions 2.0 of both BiG-SCAPE and BiG-SLiCE have been released! With significant speed and accuracy increases, as well as new interactive functionalities. www.biorxiv.org/content/10.1...

BiG-SCAPE 2.0 and BiG-SLiCE 2.0: scalable, accurate and interactive sequence clustering of metabolic gene clusters
Microbial metabolic gene clusters encode the biosynthesis or catabolism of metabolites that facilitate ecological specialization, mediate microbiome interactions and constitute a major source of medic...
www.biorxiv.org
September 1, 2025 at 10:35 PM
Now out as preprint: versions 2.0 of both BiG-SCAPE and BiG-SLiCE have been released! With significant speed and accuracy increases, as well as new interactive functionalities. www.biorxiv.org/content/10.1...
Now out as preprint: versions 2.0 of both BiG-SCAPE and BiG-SLiCE have been released! With significant speed and accuracy increases, as well as new interactive functionalities. www.biorxiv.org/content/10.1...

BiG-SCAPE 2.0 and BiG-SLiCE 2.0: scalable, accurate and interactive sequence clustering of metabolic gene clusters
Microbial metabolic gene clusters encode the biosynthesis or catabolism of metabolites that facilitate ecological specialization, mediate microbiome interactions and constitute a major source of medic...
www.biorxiv.org
September 1, 2025 at 10:35 PM
Now out as preprint: versions 2.0 of both BiG-SCAPE and BiG-SLiCE have been released! With significant speed and accuracy increases, as well as new interactive functionalities. www.biorxiv.org/content/10.1...
Reposted by Marnix Medema
Into natural product biosynthesis & tailoring enzymes? Frustrated by the lack of a dedicated resource to explore their functions? Tired of endless literature searches for reaction info? Meet the MITE database, freely available at mite.bioinformatics.nl. Preprint: doi.org/10.26434/che... (1/8)

August 21, 2025 at 10:52 AM
Into natural product biosynthesis & tailoring enzymes? Frustrated by the lack of a dedicated resource to explore their functions? Tired of endless literature searches for reaction info? Meet the MITE database, freely available at mite.bioinformatics.nl. Preprint: doi.org/10.26434/che... (1/8)
Check out the new MITE database! Kudos to @mmzdouc.bsky.social for his leadership in getting this community effort off the ground. Hopefully we can add many more data points with everyone's input!
Into natural product biosynthesis & tailoring enzymes? Frustrated by the lack of a dedicated resource to explore their functions? Tired of endless literature searches for reaction info? Meet the MITE database, freely available at mite.bioinformatics.nl. Preprint: doi.org/10.26434/che... (1/8)

August 21, 2025 at 1:07 PM
Check out the new MITE database! Kudos to @mmzdouc.bsky.social for his leadership in getting this community effort off the ground. Hopefully we can add many more data points with everyone's input!
Reposted by Marnix Medema
Finally published 🥳 We mine public transcriptome data to enhance the known diversity of iflaviruses in Lepidoptera. Virus diversity is everywhere - from the family level to the within-species level - academic.oup.com/ve/article/1... with @astridbryon.bsky.social and others #VirEvol

Insights into diversity, host range, and evolution of iflaviruses in Lepidoptera through transcriptome mining
Abstract. Insects are associated with a wide variety of diverse RNA viruses, including iflaviruses, a group of positive stranded RNA viruses that mainly in
academic.oup.com
August 14, 2025 at 1:04 PM
Finally published 🥳 We mine public transcriptome data to enhance the known diversity of iflaviruses in Lepidoptera. Virus diversity is everywhere - from the family level to the within-species level - academic.oup.com/ve/article/1... with @astridbryon.bsky.social and others #VirEvol
Reposted by Marnix Medema
Elucidating #plant #Biosynthetic pathways: @jjjvanderhooft.bsky.social @marnixmedema.bsky.social &co develop #MEANtools, an unsupervised computational workflow that integrates #MultiOmics data to predict #metabolic pathways by linking transcripts to metabolites @plosbiology.org 🧪 plos.io/4odL94g
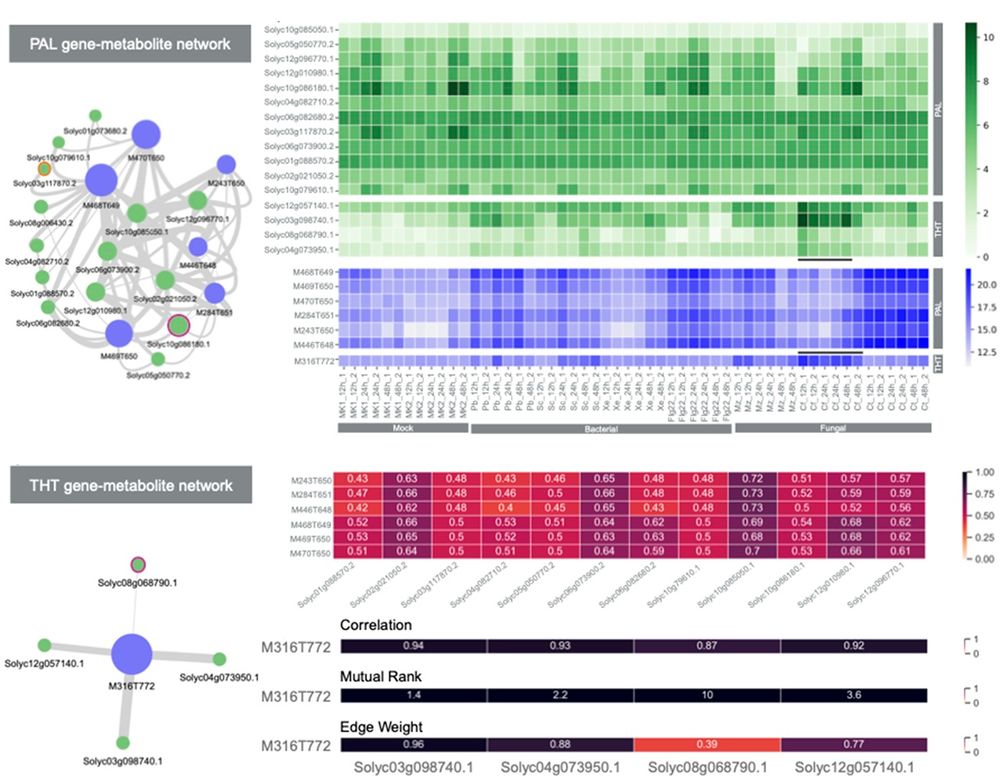
July 31, 2025 at 8:49 AM
Elucidating #plant #Biosynthetic pathways: @jjjvanderhooft.bsky.social @marnixmedema.bsky.social &co develop #MEANtools, an unsupervised computational workflow that integrates #MultiOmics data to predict #metabolic pathways by linking transcripts to metabolites @plosbiology.org 🧪 plos.io/4odL94g
Reposted by Marnix Medema
Check out the Ziemert Lab’s new YouTube channel
m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts
m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts

ZiemertLab
The Ziemert lab is interested in the evolution and distribution of bacterial secondary metabolites. These bioactive compounds are especially important in human medicine as the chemical scaffolds are t...
m.youtube.com
July 8, 2025 at 8:09 PM
Check out the Ziemert Lab’s new YouTube channel
m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts
m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts


