Computational Mass Spectrometry, Bioinformatics.
#massspec #molecularnetworking #GNPS #MassQL
https://www.cs.ucr.edu/~mingxunw/
aprecruit.ucr.edu/JPF02151

aprecruit.ucr.edu/JPF02151
metabolomics-usi.gnps2.org/dashinterfac...

metabolomics-usi.gnps2.org/dashinterfac...
We’re building a tool for repository-scale untargeted #metabolomics and #exposomics of #environmental data. To make it the best it can be, we’re looking for people willing to share high-resolution LC-MS/MS (DDA) data from #water, #soil, #sediment, and related samples.

We’re building a tool for repository-scale untargeted #metabolomics and #exposomics of #environmental data. To make it the best it can be, we’re looking for people willing to share high-resolution LC-MS/MS (DDA) data from #water, #soil, #sediment, and related samples.
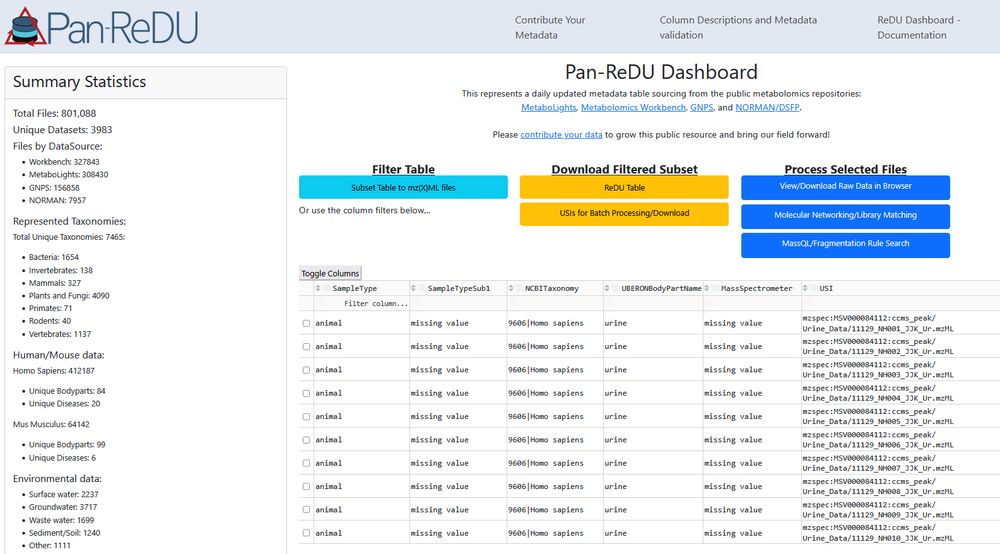
www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.cell.com/cell/fulltex...
#space #metabolomics #microbes
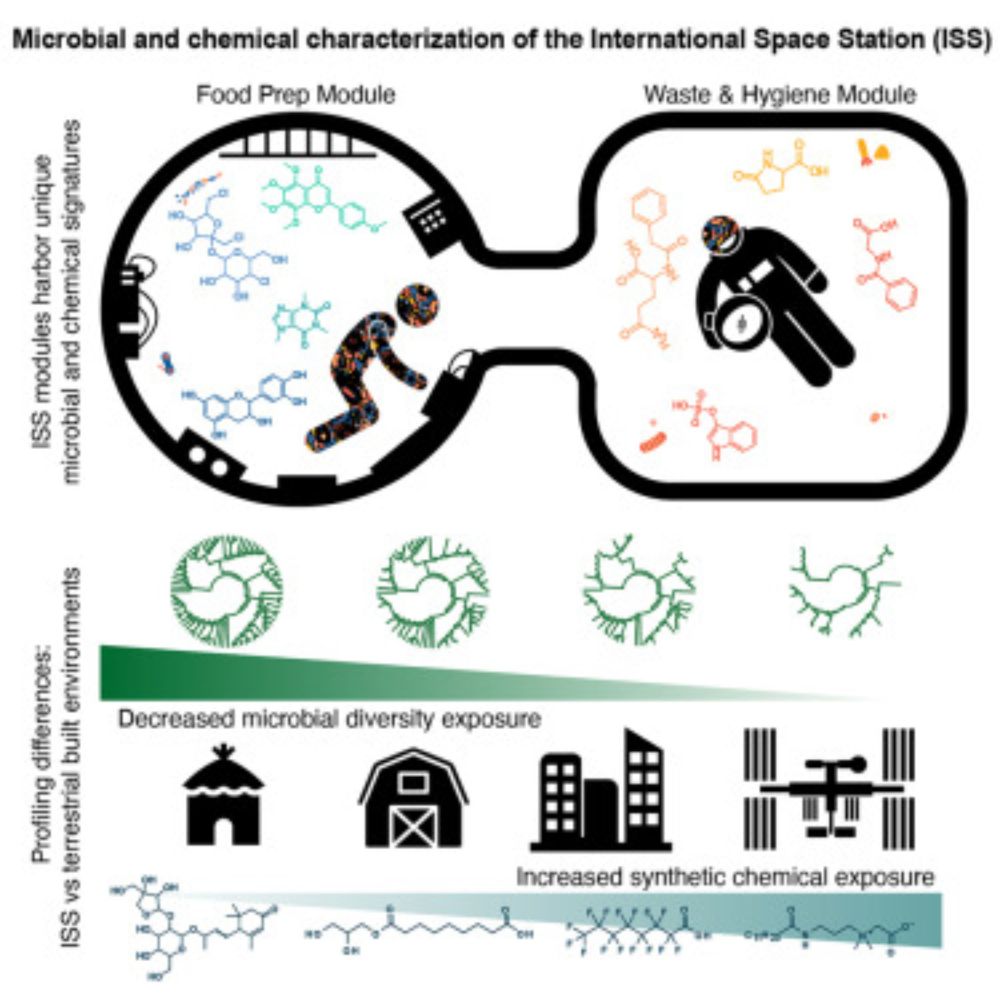
www.cell.com/cell/fulltex...
#space #metabolomics #microbes

doi.org/10.1021/acs....
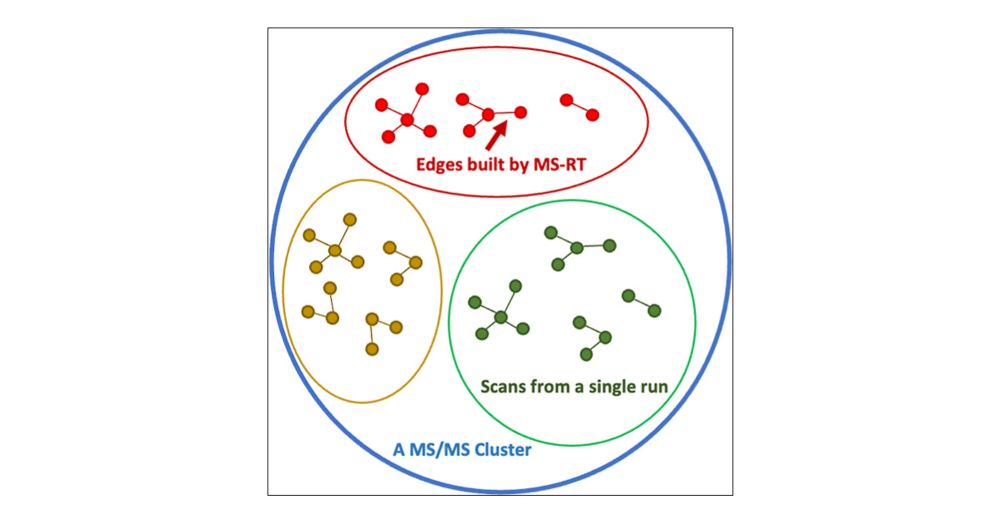
doi.org/10.1021/acs....
Don’t forget to join us for our first webinar of the year:
“Learning From Repository-Scale Untargeted Metabolomics Data”
📅 Date: 22 January
🕒 Time: 3 PM UTC
#MetabolomicsSociety #MetSoc #Metabolomics #ECR #TeamMassSpec #EMNMetSoc
On 22 January at 3pm UTC, we will host Dr Wout Bittremieux who will deliver a talk on an approach to untargeted metabolomics data annotation!
✒️ Register here for free: tinyurl.com/2v654vp9
We look forward to seeing you there 👋

Don’t forget to join us for our first webinar of the year:
“Learning From Repository-Scale Untargeted Metabolomics Data”
📅 Date: 22 January
🕒 Time: 3 PM UTC
#MetabolomicsSociety #MetSoc #Metabolomics #ECR #TeamMassSpec #EMNMetSoc





Learn about the election & meet the candidates here:
www.asms.org/about/board-...

Learn about the election & meet the candidates here:
www.asms.org/about/board-...


