Kresten Lindorff-Larsen
@lindorfflarsen.bsky.social
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰
Pinned

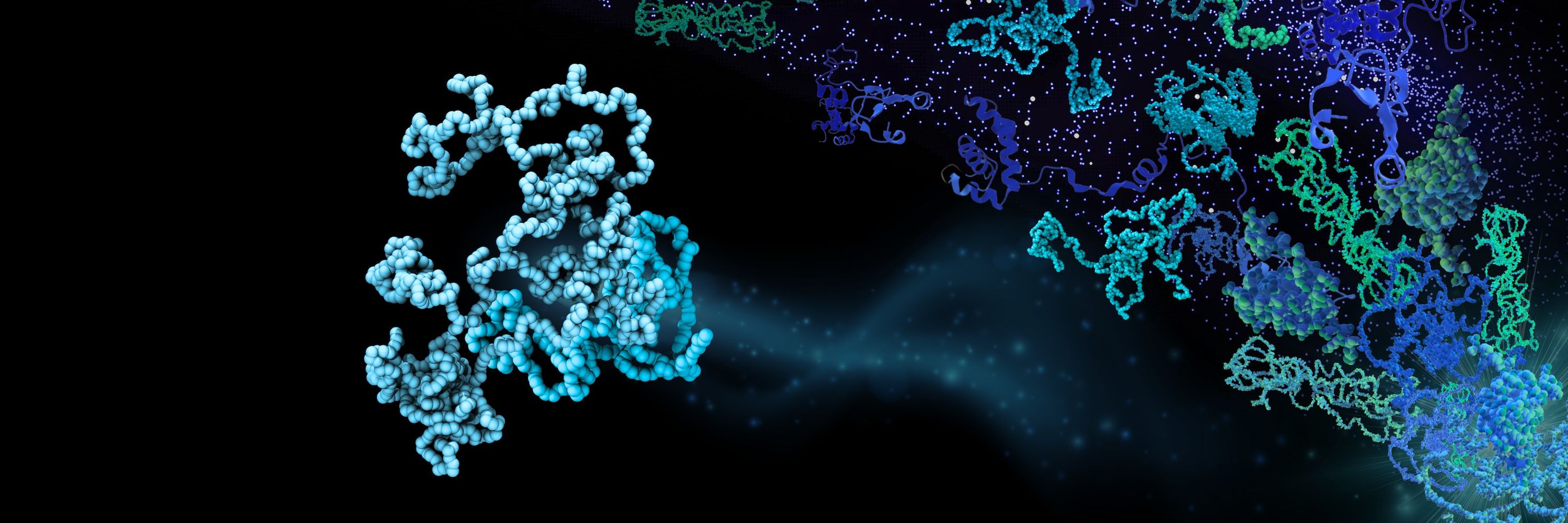
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Reposted by Kresten Lindorff-Larsen
Excited to share our latest @nature.com: How does naloxone (Narcan) stop an opioid overdose? We determined the first GDP-bound μ-opioid receptor–G protein structures and found naloxone traps a novel "latent” state, preventing GDP release and G protein activation.💊🧪 🧵👇 www.nature.com/articles/s41...
November 5, 2025 at 4:23 PM
Excited to share our latest @nature.com: How does naloxone (Narcan) stop an opioid overdose? We determined the first GDP-bound μ-opioid receptor–G protein structures and found naloxone traps a novel "latent” state, preventing GDP release and G protein activation.💊🧪 🧵👇 www.nature.com/articles/s41...
Reposted by Kresten Lindorff-Larsen
🎙️ Next up Dec 2 in VESS!
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

November 4, 2025 at 6:31 PM
🎙️ Next up Dec 2 in VESS!
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
Reposted by Kresten Lindorff-Larsen
*If you would like to be considered for a talk at this workshop, please register & submit an abstract by 7 November*
We also have a fantastic line-up of invited speakers: Matteo Degiacomi, Lucie Delemotte, Weria Pezeshkian, Mohsen Sadeghi & Ilpo Vattulainen
Registration: aias.au.dk/events/show/...
We also have a fantastic line-up of invited speakers: Matteo Degiacomi, Lucie Delemotte, Weria Pezeshkian, Mohsen Sadeghi & Ilpo Vattulainen
Registration: aias.au.dk/events/show/...
November 4, 2025 at 11:46 AM
*If you would like to be considered for a talk at this workshop, please register & submit an abstract by 7 November*
We also have a fantastic line-up of invited speakers: Matteo Degiacomi, Lucie Delemotte, Weria Pezeshkian, Mohsen Sadeghi & Ilpo Vattulainen
Registration: aias.au.dk/events/show/...
We also have a fantastic line-up of invited speakers: Matteo Degiacomi, Lucie Delemotte, Weria Pezeshkian, Mohsen Sadeghi & Ilpo Vattulainen
Registration: aias.au.dk/events/show/...
Reposted by Kresten Lindorff-Larsen
Probing the energy landscape of α-Synuclein amyloid fibril formation by systematic K-to-Q mutagenesis https://www.biorxiv.org/content/10.1101/2025.11.01.685997v1
November 3, 2025 at 10:48 PM
Probing the energy landscape of α-Synuclein amyloid fibril formation by systematic K-to-Q mutagenesis https://www.biorxiv.org/content/10.1101/2025.11.01.685997v1
Reposted by Kresten Lindorff-Larsen
Anyway, we need some joy, so here's the Egyptian foreign minister being given a Lego Pyramid by the Danish foreign minister.

November 3, 2025 at 2:35 PM
Anyway, we need some joy, so here's the Egyptian foreign minister being given a Lego Pyramid by the Danish foreign minister.
I’m excited to be speaking at the AI in Science Summit 2025 in my home town of Copenhagen #AIS25 #AI4Science
I will be speaking about how AI and biophysics can come together to study protein dynamics
I will be speaking about how AI and biophysics can come together to study protein dynamics



November 3, 2025 at 8:09 AM
I’m excited to be speaking at the AI in Science Summit 2025 in my home town of Copenhagen #AIS25 #AI4Science
I will be speaking about how AI and biophysics can come together to study protein dynamics
I will be speaking about how AI and biophysics can come together to study protein dynamics
Reposted by Kresten Lindorff-Larsen
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
CECAM - Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET data
www.cecam.org
November 1, 2025 at 9:47 AM
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Reposted by Kresten Lindorff-Larsen
Quantum computers - what are they and how will they change things?
Have a listen/watch here: @timcoulson.bsky.social and I chat with Ian Walmsley, newly appointed director of the Oxford Quantum Institute: The Science and Promise of Quantum Computing
youtu.be/sPTyvBKJruU
Have a listen/watch here: @timcoulson.bsky.social and I chat with Ian Walmsley, newly appointed director of the Oxford Quantum Institute: The Science and Promise of Quantum Computing
youtu.be/sPTyvBKJruU

Science Of The Times Ep 29_Entangled Futures: The Science and Promise of Quantum Computing
YouTube video by Science Of The Times Podcast
youtu.be
November 1, 2025 at 8:24 AM
Quantum computers - what are they and how will they change things?
Have a listen/watch here: @timcoulson.bsky.social and I chat with Ian Walmsley, newly appointed director of the Oxford Quantum Institute: The Science and Promise of Quantum Computing
youtu.be/sPTyvBKJruU
Have a listen/watch here: @timcoulson.bsky.social and I chat with Ian Walmsley, newly appointed director of the Oxford Quantum Institute: The Science and Promise of Quantum Computing
youtu.be/sPTyvBKJruU
Reposted by Kresten Lindorff-Larsen
Where Parisians post their preprints…

October 31, 2025 at 6:28 PM
Where Parisians post their preprints…
Accessible, uniform protein property prediction with a scikit-learn based toolset AIDE
Artificial Intelligence Driven protein Estimation (AIDE), enables instantiating, optimizing, and testing many zero-shot and supervised property prediction methods
doi.org/10.1093/bioi...
Artificial Intelligence Driven protein Estimation (AIDE), enables instantiating, optimizing, and testing many zero-shot and supervised property prediction methods
doi.org/10.1093/bioi...


October 29, 2025 at 7:45 AM
Accessible, uniform protein property prediction with a scikit-learn based toolset AIDE
Artificial Intelligence Driven protein Estimation (AIDE), enables instantiating, optimizing, and testing many zero-shot and supervised property prediction methods
doi.org/10.1093/bioi...
Artificial Intelligence Driven protein Estimation (AIDE), enables instantiating, optimizing, and testing many zero-shot and supervised property prediction methods
doi.org/10.1093/bioi...
Not just #YAAFC
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
October 28, 2025 at 9:54 PM
Not just #YAAFC
I attended a summer school on Spetses 🇬🇷 just as I was beginning my PhD. Looks like I will be going again in 2026 with "Lost in Integration vol. 2". Details to follow.

October 28, 2025 at 9:22 PM
I attended a summer school on Spetses 🇬🇷 just as I was beginning my PhD. Looks like I will be going again in 2026 with "Lost in Integration vol. 2". Details to follow.
New review on experimental datasets that can be used to benchmark protein force fields. And if that doesn’t tickle your fancy, the data can also be used to benchmark machine learning models for biomolecular structure and dynamics.
In the latest @livecomsjournal.bsky.social
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem

October 28, 2025 at 3:51 PM
New review on experimental datasets that can be used to benchmark protein force fields. And if that doesn’t tickle your fancy, the data can also be used to benchmark machine learning models for biomolecular structure and dynamics.
Reposted by Kresten Lindorff-Larsen
Looks like OpenFold3 has been formally released in a public "preview". Not quite on parity with AlphaFold3 on a few benchmarks shown, in particular for antibody interactions. All info on the github link. I am sure we will hear more about this from the developers github.com/aqlaboratory...

GitHub - aqlaboratory/openfold-3: OpenFold3: A fully open source biomolecular structure prediction model based on AlphaFold3
OpenFold3: A fully open source biomolecular structure prediction model based on AlphaFold3 - aqlaboratory/openfold-3
github.com
October 28, 2025 at 9:30 AM
Looks like OpenFold3 has been formally released in a public "preview". Not quite on parity with AlphaFold3 on a few benchmarks shown, in particular for antibody interactions. All info on the github link. I am sure we will hear more about this from the developers github.com/aqlaboratory...
Reposted by Kresten Lindorff-Larsen
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
October 27, 2025 at 7:54 AM
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
Just because this came up a few times recently
If you need to compare two or more ensembles (like RMSD for structures) we developed algorithms and code (ENCORE) to do just that
Similarity Measures for Protein Ensembles
journals.plos.org/plosone/arti...
ENCORE
journals.plos.org/ploscompbiol...
If you need to compare two or more ensembles (like RMSD for structures) we developed algorithms and code (ENCORE) to do just that
Similarity Measures for Protein Ensembles
journals.plos.org/plosone/arti...
ENCORE
journals.plos.org/ploscompbiol...

October 27, 2025 at 8:00 AM
Just because this came up a few times recently
If you need to compare two or more ensembles (like RMSD for structures) we developed algorithms and code (ENCORE) to do just that
Similarity Measures for Protein Ensembles
journals.plos.org/plosone/arti...
ENCORE
journals.plos.org/ploscompbiol...
If you need to compare two or more ensembles (like RMSD for structures) we developed algorithms and code (ENCORE) to do just that
Similarity Measures for Protein Ensembles
journals.plos.org/plosone/arti...
ENCORE
journals.plos.org/ploscompbiol...
Reposted by Kresten Lindorff-Larsen
Looking forward to hosting Kresten Lindorff-Larsen's talk tomorrow.
I am looking forward to seeing ISTA's main lecture hall of full, and hearing the latest developments of tools to predict, design and study intrinsically disordered proteins.
@istaresearch.bsky.social
I am looking forward to seeing ISTA's main lecture hall of full, and hearing the latest developments of tools to predict, design and study intrinsically disordered proteins.
@istaresearch.bsky.social
Looking forward to visit colleagues in Vienna and Graz 🇦🇹
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
October 26, 2025 at 6:38 PM
Looking forward to hosting Kresten Lindorff-Larsen's talk tomorrow.
I am looking forward to seeing ISTA's main lecture hall of full, and hearing the latest developments of tools to predict, design and study intrinsically disordered proteins.
@istaresearch.bsky.social
I am looking forward to seeing ISTA's main lecture hall of full, and hearing the latest developments of tools to predict, design and study intrinsically disordered proteins.
@istaresearch.bsky.social
Looking forward to visit colleagues in Vienna and Graz 🇦🇹
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
October 26, 2025 at 6:41 AM
Looking forward to visit colleagues in Vienna and Graz 🇦🇹
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
If you’re in Vienna I’m speaking on:
Prediction and Design of Intrinsically Disordered Proteins and Condensates (27/10 ISTA)
&
Understanding the Effects of Missense Variants Using Analyses of Protein Stability and Conservation (30/10 VBC)
Reposted by Kresten Lindorff-Larsen
Remember, everyone, the best publication metric is the one that makes *your* publication record look the best.
My first thought was that Google took a page directly from the @drugmonkey.bsky.social blog post from awhile back on the h-index gamification.
www.nature.com/articles/d41...
www.nature.com/articles/d41...

Google Scholar tool gives extra credit to first and last authors
Researchers welcome the initiative, but say it doesn’t go far enough to capture the nuance of researcher productivity and impact.
www.nature.com
October 25, 2025 at 4:00 PM
Remember, everyone, the best publication metric is the one that makes *your* publication record look the best.
Reposted by Kresten Lindorff-Larsen
A machine learning method for calculating highly localized protein stabilities https://www.biorxiv.org/content/10.1101/2025.10.21.683809v1
October 23, 2025 at 3:49 PM
A machine learning method for calculating highly localized protein stabilities https://www.biorxiv.org/content/10.1101/2025.10.21.683809v1
Reposted by Kresten Lindorff-Larsen
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
October 22, 2025 at 2:55 PM
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
Reposted by Kresten Lindorff-Larsen
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

October 20, 2025 at 11:26 AM
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Reposted by Kresten Lindorff-Larsen
We're excited to present LeaVS, a method to scale up learning for protein function models. It is based on the co-design of wet lab experiments and in silico training.

October 21, 2025 at 2:38 PM
We're excited to present LeaVS, a method to scale up learning for protein function models. It is based on the co-design of wet lab experiments and in silico training.
Reposted by Kresten Lindorff-Larsen
Submit your BPS T&C subgroup nominations::!!
Hello @biophysicalsoc.bsky.social Theory & Computation Subgroup! Please consider volunteering as an officer for our subgroup for the 2026-2028 cycle. Submit your nomination(s) by November 17th.
Looking forward to seeing you next Annual Meeting in San Francisco next year!!
Looking forward to seeing you next Annual Meeting in San Francisco next year!!

October 21, 2025 at 2:20 PM
Submit your BPS T&C subgroup nominations::!!
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

October 20, 2025 at 11:26 AM
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

