
1️⃣ Stephan Riesenberg (Group Leader, Max Planck Leipzig): CRISPR-mediated generation of genetic variants for functional analysis.
2️⃣ Yuriy Baglaenko @baglaenkolab.bsky.social: Causal variants with CRISPR editing in primary human cells www.varianteffect.org/seminar-seri...

1️⃣ Stephan Riesenberg (Group Leader, Max Planck Leipzig): CRISPR-mediated generation of genetic variants for functional analysis.
2️⃣ Yuriy Baglaenko @baglaenkolab.bsky.social: Causal variants with CRISPR editing in primary human cells www.varianteffect.org/seminar-seri...
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
🗓️ Early bird registration & abstract submissions close Nov 2, 2025.
www.mss2026.org
🗓️ Early bird registration & abstract submissions close Nov 2, 2025.
www.mss2026.org
Population genetics × variant effects:
🧬 Nikhil Milind (Stanford) on gene dosage and complex traits @nikhilmilind.dev
🧬 Leslie Smith (U Florida) on equitable ML in cancer genomics
www.varianteffect.org/seminar-seri...
@varianteffect.bsky.social

Population genetics × variant effects:
🧬 Nikhil Milind (Stanford) on gene dosage and complex traits @nikhilmilind.dev
🧬 Leslie Smith (U Florida) on equitable ML in cancer genomics
www.varianteffect.org/seminar-seri...
@varianteffect.bsky.social
docs.google.com/document/d/1...

docs.google.com/document/d/1...
First speaker: Shelby Hemker (Dr. Jacob Kitzman Lab, University of Michigan)
Second speaker: Karl Romanowicz (Dr. Calin Plesa Lab, University of Oregon) @kroman.bsky.social
Link: www.varianteffect.org/seminar-seri...

First speaker: Shelby Hemker (Dr. Jacob Kitzman Lab, University of Michigan)
Second speaker: Karl Romanowicz (Dr. Calin Plesa Lab, University of Oregon) @kroman.bsky.social
Link: www.varianteffect.org/seminar-seri...
We present a new type of cell fitness assay that allows you to both quantify and explain differences across human donors in cell proliferation and sensitivity to environmental toxicants.
We present a new type of cell fitness assay that allows you to both quantify and explain differences across human donors in cell proliferation and sensitivity to environmental toxicants.
bsky.app/profile/bioi...
Full article available: https://doi.org/10.1093/bioadv/vbaf218

bsky.app/profile/bioi...
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)

Details below
careersearch.stanford.edu/jobs/computa...
Plz RT


Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
ℹ️ www.varianteffect.org/previous-sem... 📺 www.youtube.com/playlist?lis...
#Genomics #Seminar #EarlyCareerResearchers #ScientificSeminar #PrecisionMedicine

ℹ️ www.varianteffect.org/previous-sem... 📺 www.youtube.com/playlist?lis...
#Genomics #Seminar #EarlyCareerResearchers #ScientificSeminar #PrecisionMedicine
bsky.app/profile/jing...
Meet Cosmos, our new statistical framework for causal inference in multi-phenotype DMS.
www.biorxiv.org/content/10.1...
[1/n]

bsky.app/profile/jing...
Meet Cosmos, our new statistical framework for causal inference in multi-phenotype DMS.
www.biorxiv.org/content/10.1...
[1/n]

Meet Cosmos, our new statistical framework for causal inference in multi-phenotype DMS.
www.biorxiv.org/content/10.1...
[1/n]

probgen2026.github.io
Please help spread the news.
probgen2026.github.io
Please help spread the news.



I think my favorite story from the supplement is how impactful normalization can be in this context.
bsky.app/profile/jero...
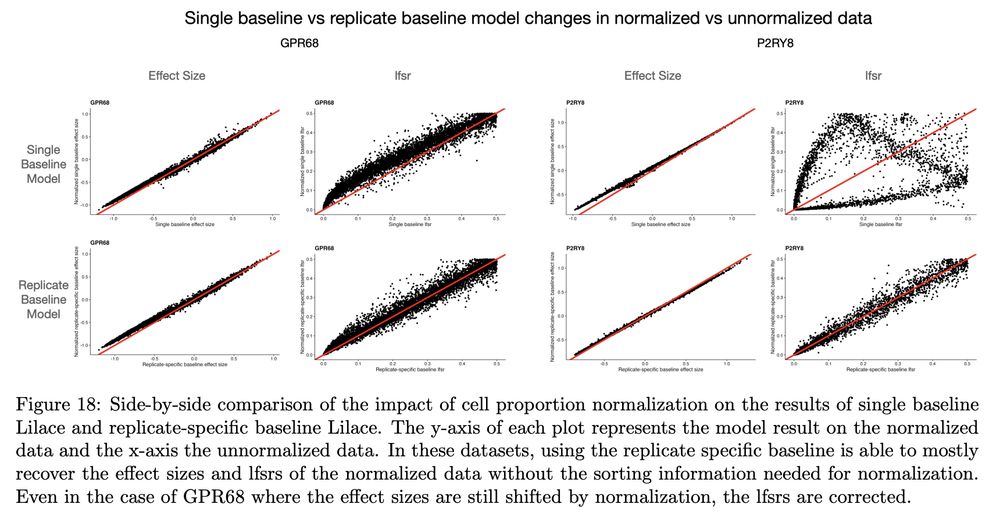
I think my favorite story from the supplement is how impactful normalization can be in this context.
bsky.app/profile/jero...


