Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
Population genetics × variant effects:
🧬 Nikhil Milind (Stanford) on gene dosage and complex traits @nikhilmilind.dev
🧬 Leslie Smith (U Florida) on equitable ML in cancer genomics
www.varianteffect.org/seminar-seri...
@varianteffect.bsky.social

Population genetics × variant effects:
🧬 Nikhil Milind (Stanford) on gene dosage and complex traits @nikhilmilind.dev
🧬 Leslie Smith (U Florida) on equitable ML in cancer genomics
www.varianteffect.org/seminar-seri...
@varianteffect.bsky.social
First speaker: Shelby Hemker (Dr. Jacob Kitzman Lab, University of Michigan)
Second speaker: Karl Romanowicz (Dr. Calin Plesa Lab, University of Oregon) @kroman.bsky.social
Link: www.varianteffect.org/seminar-seri...

First speaker: Shelby Hemker (Dr. Jacob Kitzman Lab, University of Michigan)
Second speaker: Karl Romanowicz (Dr. Calin Plesa Lab, University of Oregon) @kroman.bsky.social
Link: www.varianteffect.org/seminar-seri...
Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]

Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
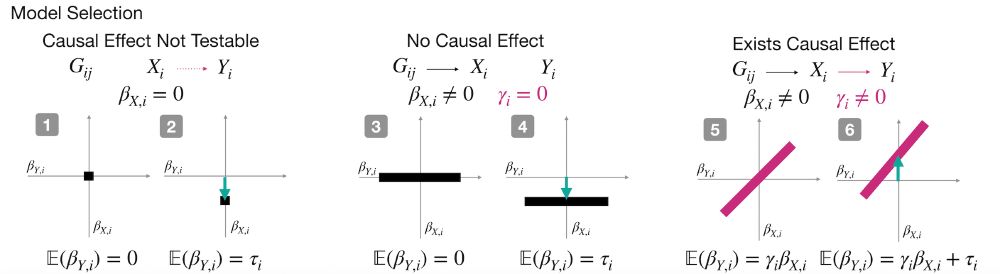
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
No need for detailed biophysical models—just stats and data.
[3/n]

No need for detailed biophysical models—just stats and data.
[3/n]
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]
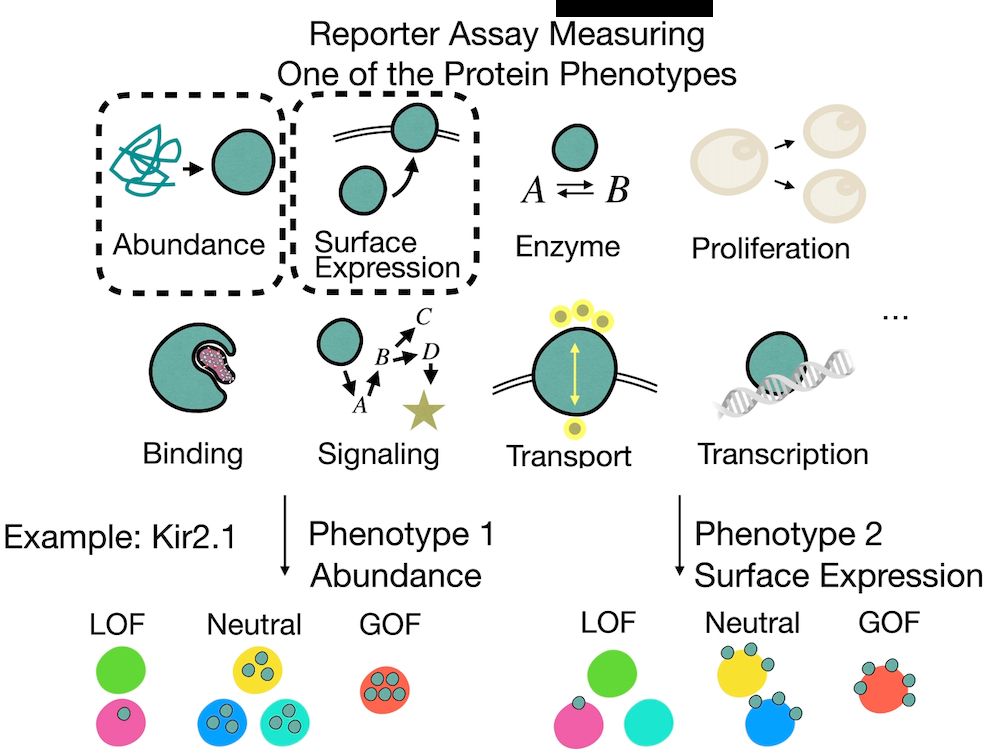
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]