Jingyou Rao
@jingyour.bsky.social
Incoming Postdoc @UCSF wcoyotelab.com | PhD in Computer Science @UCLA protein epistasis and mutational scanning
We applied Cosmos to 3 DMS datasets:
Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]
Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]

August 4, 2025 at 3:10 PM
We applied Cosmos to 3 DMS datasets:
Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]
Kir2.1: abundance → surface expression
PDZ3: abundance → CRIPT binding
KRAS: abundance → RAF1_RBD binding
Cosmos clearly separates direct binding residues from those with indirect effects.
[6/n]
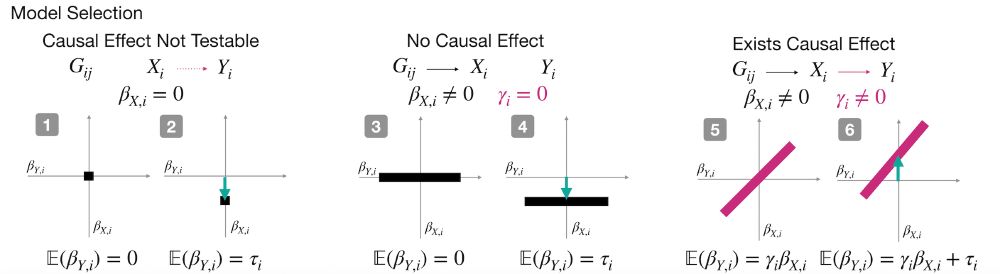
How does it work?
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
August 4, 2025 at 3:10 PM
How does it work?
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects.
[5/n]
Cosmos answers three key questions:
1️⃣ Is there a causal link between the phenotypes?
2️⃣ How strong is it?
3️⃣ What would the downstream phenotype look like if we “normalized” the upstream one (i.e., counterfactual inference)?
[4/n]
1️⃣ Is there a causal link between the phenotypes?
2️⃣ How strong is it?
3️⃣ What would the downstream phenotype look like if we “normalized” the upstream one (i.e., counterfactual inference)?
[4/n]
August 4, 2025 at 3:09 PM
Cosmos answers three key questions:
1️⃣ Is there a causal link between the phenotypes?
2️⃣ How strong is it?
3️⃣ What would the downstream phenotype look like if we “normalized” the upstream one (i.e., counterfactual inference)?
[4/n]
1️⃣ Is there a causal link between the phenotypes?
2️⃣ How strong is it?
3️⃣ What would the downstream phenotype look like if we “normalized” the upstream one (i.e., counterfactual inference)?
[4/n]
Cosmos is a Bayesian framework that performs residue-level causal inference to decouple how mutations influence upstream vs downstream functions.
No need for detailed biophysical models—just stats and data.
[3/n]
No need for detailed biophysical models—just stats and data.
[3/n]

August 4, 2025 at 3:09 PM
Cosmos is a Bayesian framework that performs residue-level causal inference to decouple how mutations influence upstream vs downstream functions.
No need for detailed biophysical models—just stats and data.
[3/n]
No need for detailed biophysical models—just stats and data.
[3/n]
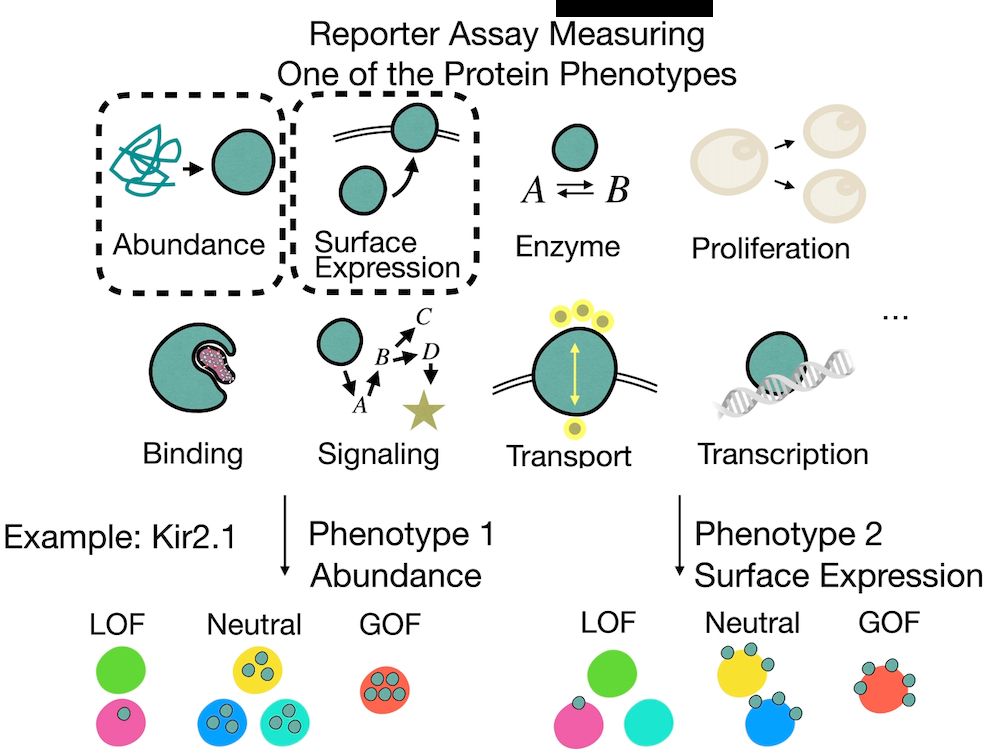
Multi-phenotype DMS experiments are revealing how mutations impact different protein functions.
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]
August 4, 2025 at 3:09 PM
Multi-phenotype DMS experiments are revealing how mutations impact different protein functions.
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]
But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance.
So how do we tell what’s direct vs indirect?
[2/n]