
#teammassspec
open and reproducible science
postdoc at SCI MUNI (CZ)
climbing, hiking, swing dancing!
No more settling for “object of type `builtin` is not subsettable”
It even looks to see what packages are loaded!
github.com/rdboyes/errbud

No more settling for “object of type `builtin` is not subsettable”
It even looks to see what packages are loaded!
github.com/rdboyes/errbud
www.nature.com/articles/s41...
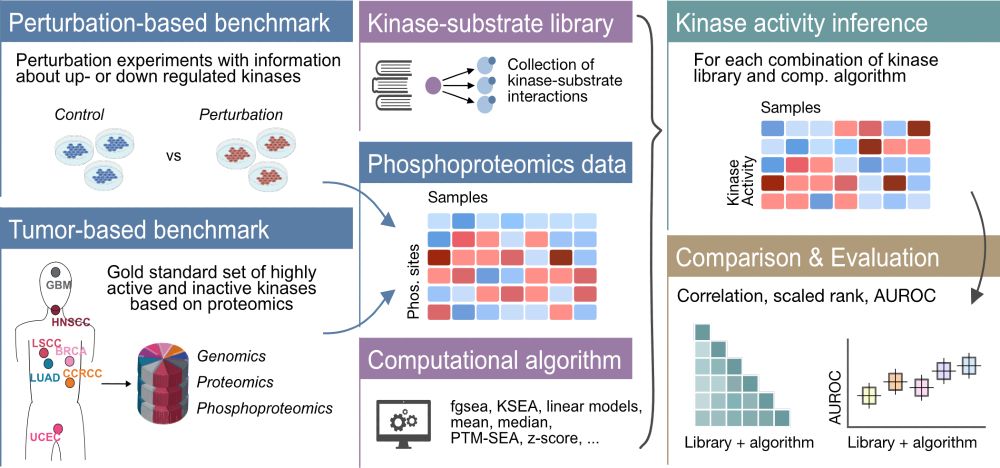
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵


www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵

🧵👇
www.biorxiv.org/content/10.1...

🧵👇
www.biorxiv.org/content/10.1...
out in @cellcellpress.bsky.social: doi.org/nzwz
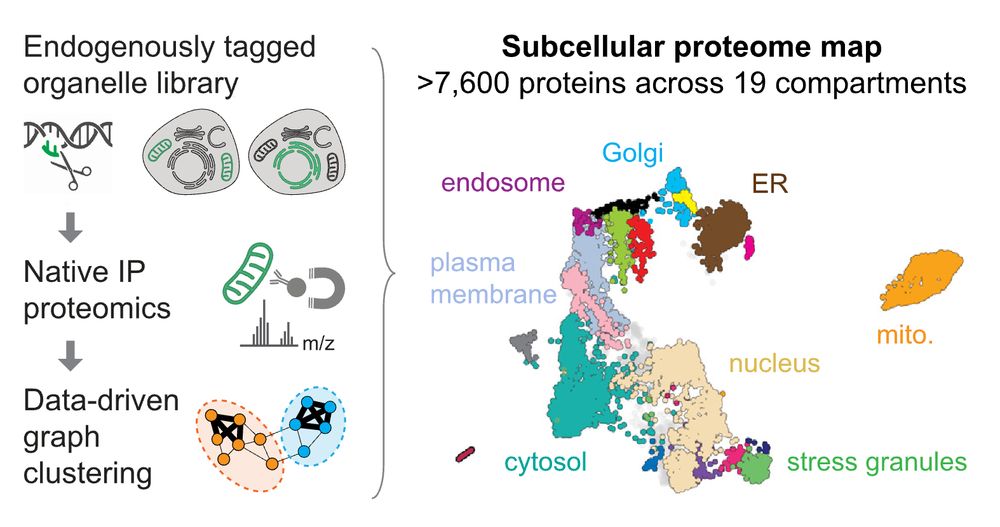
We created a spatial map of human cells using organelle immunoprecipitation at scale.
Locate your fav protein among the 7,600 we mapped across 19 subcellular compartments: organelles.sf.czbiohub.org/
1/3 🧪
out in @cellcellpress.bsky.social: doi.org/nzwz
We created a spatial map of human cells using organelle immunoprecipitation at scale.
Locate your fav protein among the 7,600 we mapped across 19 subcellular compartments: organelles.sf.czbiohub.org/
1/3 🧪
bsky.app/profile/brod...
cran.r-project.org/web/packages...
Thanks to the many contributors for this release (credited in the DESCRIPTION file)
Here is a sneak peak of what awaits you:
1/4
bsky.app/profile/brod...



For a course I teach, I am collecting papers on: cell/dev bio, microbio, neuro, maths/stats/CS/chem/phys applied to biology.
Students use one paper as the basis for a "research grant application" that they will write. 1/2
For a course I teach, I am collecting papers on: cell/dev bio, microbio, neuro, maths/stats/CS/chem/phys applied to biology.
Students use one paper as the basis for a "research grant application" that they will write. 1/2
In our latest paper, we advocate for improving software quality in bioinformatics through teamwork.
academic.oup.com/bioinformati...

In our latest paper, we advocate for improving software quality in bioinformatics through teamwork.
academic.oup.com/bioinformati...
This series of lectures may be helpful.
We aimed to make them short & accessible.
Enjoy watching & learning!
youtube.com/playlist?lis...

This series of lectures may be helpful.
We aimed to make them short & accessible.
Enjoy watching & learning!
youtube.com/playlist?lis...

