Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
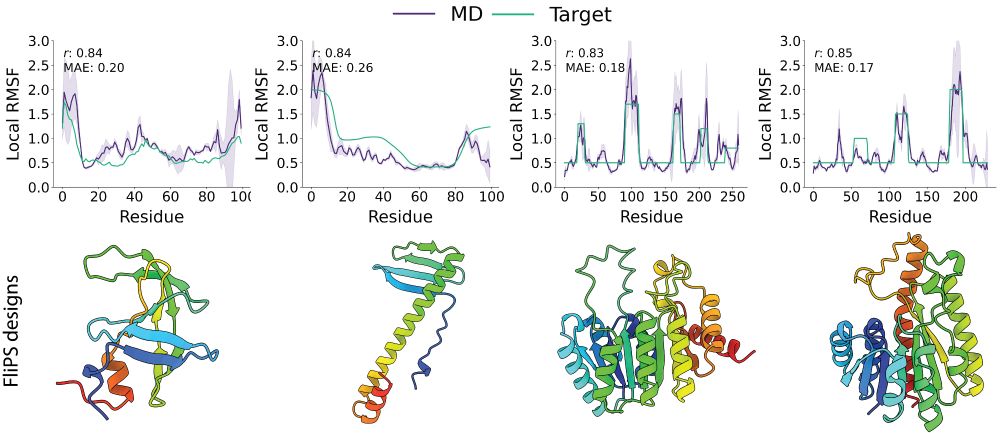
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT


Analyzes multimodal protein language models, identifying & addressing fidelity loss from 3D structure tokenization through enhanced modeling, architectures & data.




Analyzes multimodal protein language models, identifying & addressing fidelity loss from 3D structure tokenization through enhanced modeling, architectures & data.
www.biorxiv.org/content/10.1...



www.biorxiv.org/content/10.1...
Designs protein seqs w/ flow matching on compressed embeddings from protein LMs to improve efficiency & reduce training costs.




Designs protein seqs w/ flow matching on compressed embeddings from protein LMs to improve efficiency & reduce training costs.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Lyra:
Models sequence-to-function relationships using epistasis with state space models and gated convolutions, for efficient biological sequence modeling.




Lyra:
Models sequence-to-function relationships using epistasis with state space models and gated convolutions, for efficient biological sequence modeling.
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
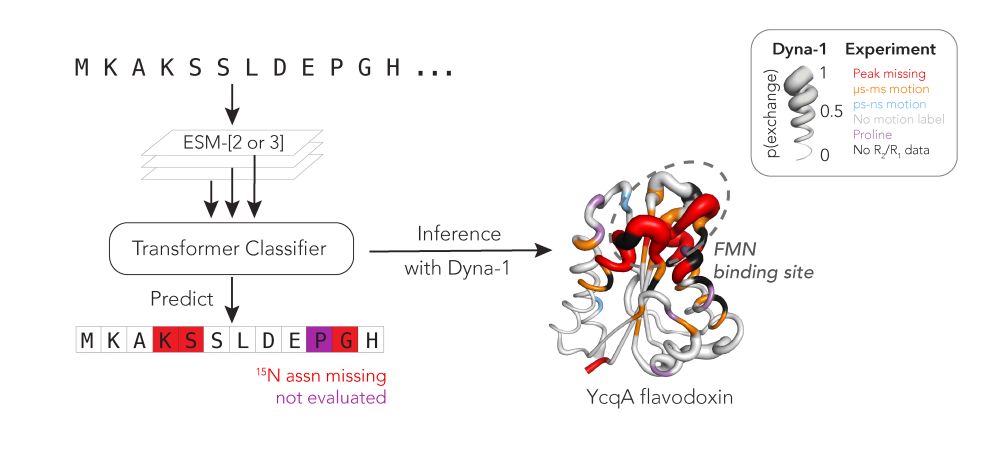
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
Open to all, with a remote keynote by Nobel laureate David Baker.
More info coming soon — it’s going to be amazing! Hope to see you there!

Open to all, with a remote keynote by Nobel laureate David Baker.
More info coming soon — it’s going to be amazing! Hope to see you there!
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and will feature an amazing list of speakers 👨🔬👩🔬👩💻👨💻
Submit your application via:
benzon-foundation.dk/benzon-sympo...

Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and will feature an amazing list of speakers 👨🔬👩🔬👩💻👨💻
Submit your application via:
benzon-foundation.dk/benzon-sympo...
Uses fragments to represent proteins as sets/graphs. Reduces dimensionality, preserves function, and aids in protein design tasks.




Uses fragments to represent proteins as sets/graphs. Reduces dimensionality, preserves function, and aids in protein design tasks.

Paper:
www.nature.com/articles/s44...
Software:
github.com/xuhuihuang/f...

Paper:
www.nature.com/articles/s44...
Software:
github.com/xuhuihuang/f...
We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv

We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv
💻 github.com/VarunUllanat...
🧵⬇️
💻 github.com/VarunUllanat...
🧵⬇️
github.com/pentalpha/pr...

github.com/pentalpha/pr...
Currently, the following models are available:
- SpliceAI
- Pangolin
- SpliceTransformer
- Borzoi
Happy to get feedback!
Currently, the following models are available:
- SpliceAI
- Pangolin
- SpliceTransformer
- Borzoi
Happy to get feedback!


Compare that to the Nobel Prize winning AI model behind AlphaFold for predictive protein modeling.
Compare that to the Nobel Prize winning AI model behind AlphaFold for predictive protein modeling.
Add it to the list
Add it to the list


