
Creator of @splicingnews.bsky.social
We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K
Mafalda, @cwjpugh.bsky.social and I will be in San Diego next week for NeurIPS -- happy to chat variant effect prediction (or just say hi).
“From Likelihood to Fitness: Improving Variant Effect Prediction in Protein and Genome Language Models”
openreview.net/pdf/a151f62e...
Mafalda, @cwjpugh.bsky.social and I will be in San Diego next week for NeurIPS -- happy to chat variant effect prediction (or just say hi).
“From Likelihood to Fitness: Improving Variant Effect Prediction in Protein and Genome Language Models”
openreview.net/pdf/a151f62e...

Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

Here’s the peer-reviewed version of our paper on how we can measure changes in splicing factor activity in virtually any single-cell dataset: doi.org/10.1093/nar/...

Here’s the peer-reviewed version of our paper on how we can measure changes in splicing factor activity in virtually any single-cell dataset: doi.org/10.1093/nar/...
Join us to develop deep generative models of cross-species data to tackle open questions in disease genetics.
www.crg.eu/en/content/t...
www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...
BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`
BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
GeneticScores.org is a new platform that enables secure, cloud-based calculation of polygenic scores to make genomic risk prediction more accessible.
www.ebi.ac.uk/about/news/u...
🖥️🧬

GeneticScores.org is a new platform that enables secure, cloud-based calculation of polygenic scores to make genomic risk prediction more accessible.
www.ebi.ac.uk/about/news/u...
🖥️🧬
This made me wonder how science would be made nowadays out of the journal publishing context. Would we try to answer different questions?
This made me wonder how science would be made nowadays out of the journal publishing context. Would we try to answer different questions?
www.biorxiv.org/content/10.1...





Here’s our approach: bsky.app/profile/bior...
Here’s our approach: bsky.app/profile/bior...
Currently, the following models are available:
- SpliceAI
- Pangolin
- SpliceTransformer
- Borzoi
Happy to get feedback!
Currently, the following models are available:
- SpliceAI
- Pangolin
- SpliceTransformer
- Borzoi
Happy to get feedback!
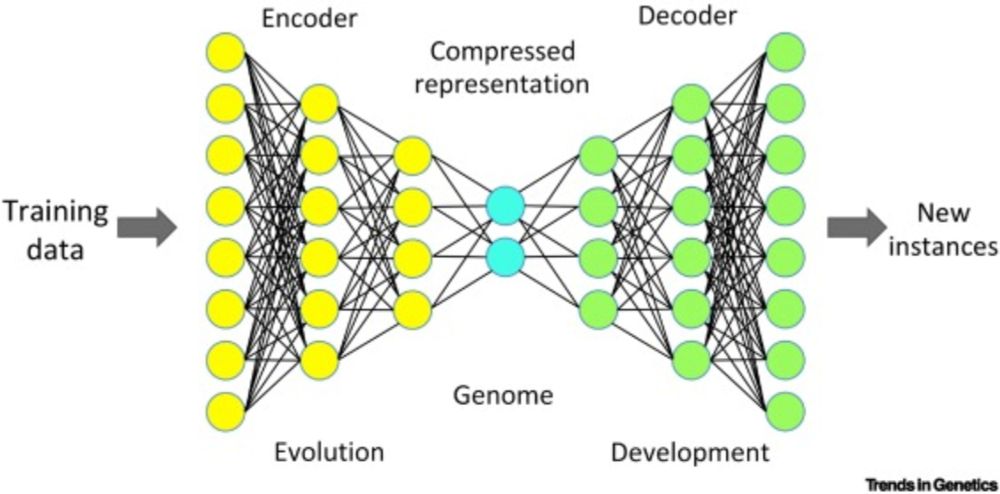

www.science.org/doi/10.1126/...
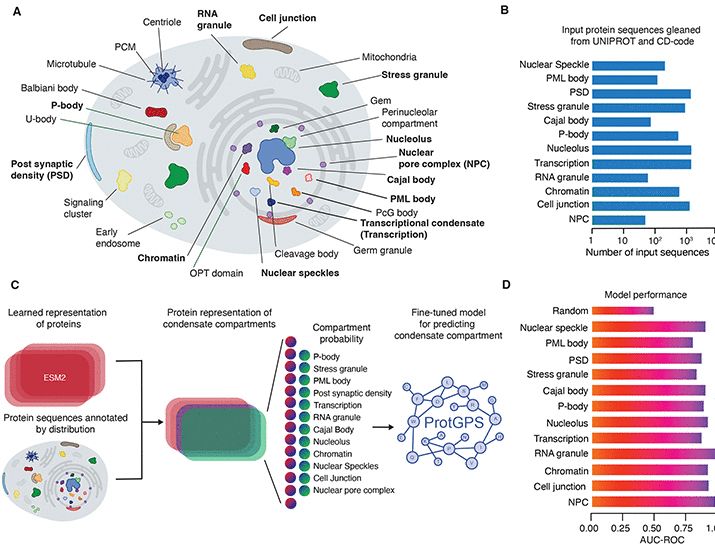
www.science.org/doi/10.1126/...

Analyzes 3D genome data with a U-shaped network & axial attention to identify loops/structures. Utilizes pretraining, for universal detection.




Analyzes 3D genome data with a U-shaped network & axial attention to identify loops/structures. Utilizes pretraining, for universal detection.
Novel model, MethylQUEEN, learns methylation states from DNA using a transformer, inferring tissue origin, gene expression and key regulatory sites.




Novel model, MethylQUEEN, learns methylation states from DNA using a transformer, inferring tissue origin, gene expression and key regulatory sites.
PAAC clustering: agglo,k-means,GMM best; spectral poor. Some annot.






