Investigator @juliazeitlinger.bsky.social is featured by @asbmb.bsky.social for her pioneering work using AI to decode the genome’s hidden grammar.
📖 Read more: bit.ly/4os5H8E

Check our latest preprint, led by @mmasoura.bsky.social, to find out!
www.biorxiv.org/content/10.1...

Check our latest preprint, led by @mmasoura.bsky.social, to find out!
www.biorxiv.org/content/10.1...


What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/

What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/

The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you





#AI tools are helping scientists, including Investigator @juliazeitlinger.bsky.social , decipher how non-coding #DNA shapes health and disease. 🧬
📖 Read more @nature.com: go.nature.com/4mLGGEl

#AI tools are helping scientists, including Investigator @juliazeitlinger.bsky.social , decipher how non-coding #DNA shapes health and disease. 🧬
📖 Read more @nature.com: go.nature.com/4mLGGEl

Register: bit.ly/3GgsQtC

Register: bit.ly/3GgsQtC
genomebiology.biomedcentral.com/articles/10....
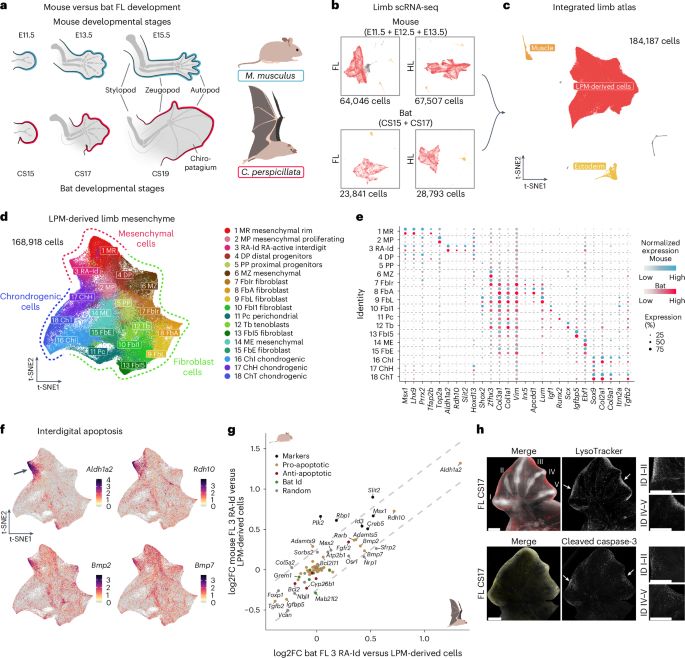
Turns out they reutilize existing gene programs 🔧
Find out more in our paper @natecoevo.nature.com in collaboration with @fany-real.bsky.social and @stemundi.bsky.social labs
www.nature.com/articles/s41...
Turns out they reutilize existing gene programs 🔧
Find out more in our paper @natecoevo.nature.com in collaboration with @fany-real.bsky.social and @stemundi.bsky.social labs
www.nature.com/articles/s41...
www.csh-asia.org?content/2743
www.csh-asia.org?content/2743

📍Scenes from check-in as the #science starts to unfold.




📍Scenes from check-in as the #science starts to unfold.
Thanks to Boyang (ex-PD) and Vivian (staff scientist) from my lab who helped train & interpret ChromBPNet models.
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
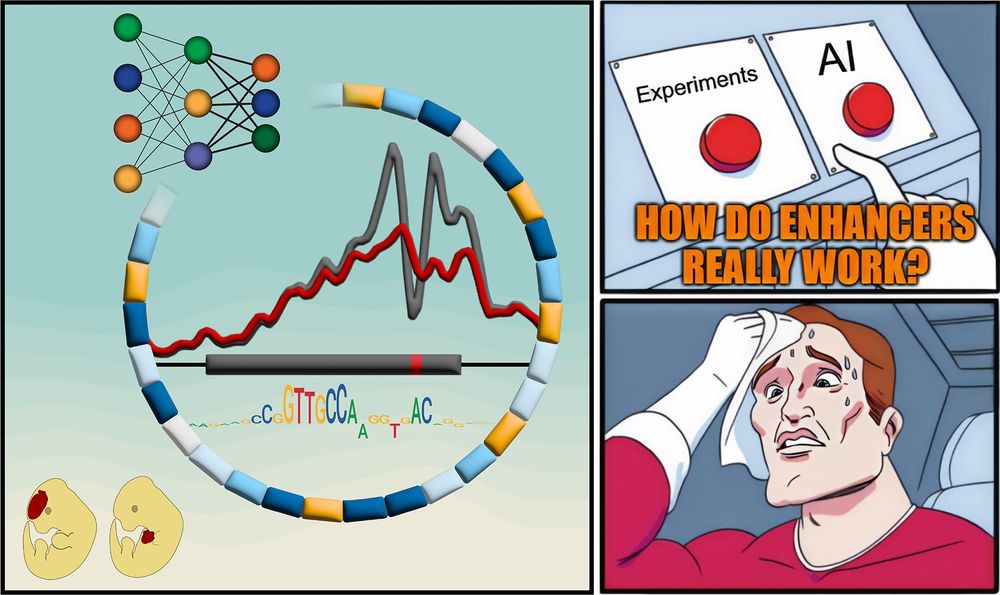
Thanks to Boyang (ex-PD) and Vivian (staff scientist) from my lab who helped train & interpret ChromBPNet models.
BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`
BPNet/ChromBPNet are powerful models for understanding regulatory genomics from @anshulkundaje.bsky.social's group, and now it's way easier to go from raw data to trained models and analysis + results in PyTorch
Try it out with `pip install bpnet-lite`